













| General Information: |
|
| Name(s) found: |
alh-10 /
CE06980
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 506 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IEA cytoplasm [IEA |
| Biological Process: |
cellular aldehyde metabolic process
[IEA metabolic process [IEA 10-formyltetrahydrofolate catabolic process [IEA proline biosynthetic process [IEA valine metabolic process [IEA betaine biosynthetic process [IEA one-carbon metabolic process [IEA |
| Molecular Function: |
1-pyrroline-5-carboxylate dehydrogenase activity
[IEA betaine-aldehyde dehydrogenase activity [IEA oxidoreductase activity [IEA aldehyde dehydrogenase [NAD(P)+] activity [IEA methylmalonate-semialdehyde dehydrogenase (acylating) activity [IEA formyltetrahydrofolate dehydrogenase activity [IEA oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASLNYREEF KNILQKLIDS PNEQHDPLTN FIGNEFARSE KLMDSVNPST GKPWIKIPDG 60
61 TAREVDQAVE AAKEAFKTWK KTTVQQRSAL LNKVANLIEE FNDDIAILES RDQGKPIGLA 120
121 KVMDIPRCVQ NFRDFANAAL YSLSTSTILE QPTGKFVNYV KNDPVGVAGL ISPWNLPLYL 180
181 LSFKLAPALV AGNTVVCKPS EMTSVTAWVL MHAFKLVGFP PGVVNMVIGE GKSAGQRLVD 240
241 HVDVPLISFT GSTVIGKKIQ EDGAKLNKKV SLEMGGKNPG IVYSNYRKSD IASIARSSFL 300
301 NQGEICLCTS RLFVQKPIFA DFVKSYVEEA KKFTVGDPTT QVQIGAMNSK VHYEKVKSYI 360
361 ELAKKEGADI LCGGVTTIQN GCENGYFITP TVIVGLHDAS KVMTDEIFGP VVCITPFDTA 420
421 EEVIERANST PYGLSATVWS SDSDELLNTA NELRAGTVWC NTWLARDLSM PFGGCKQSGN 480
481 GREGLHDSLH FYSDAKTVCV NIAAKY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
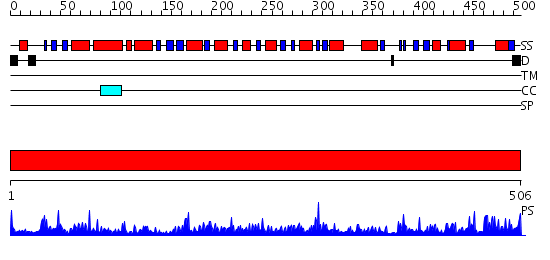
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..506] | 171.0 | Aldehyde reductase (dehydrogenase), ALDH |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)