













| General Information: |
|
| Name(s) found: |
sir-2.1 /
CE06302
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 607 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromatin silencing complex
[IEA |
| Biological Process: |
determination of adult lifespan
[IMP protein amino acid deacetylation [IDA regulation of transcription, DNA-dependent [IEA chromatin silencing [IEA |
| Molecular Function: |
DNA binding
[IEA deacetylase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRDSGNDSE VAVTHGEVQE ITEENPEIGS MHITQETDIS DAPETNTDSS RQRTESTTSV 60
61 SSESWQNNDE MMSNLRRAQR LLDDGATPLQ IIQQIFPDFN ASRIATMSEN AHFAILSDLL 120
121 ERAPVRQKLT NYNSLADAVE LFKTKKHILV LTGAGVSVSC GIPDFRSKDG IYARLRSEFP 180
181 DLPDPTAMFD IRYFRENPAP FYNFAREIFP GQFVPSVSHR FIKELETSGR LLRNYTQNID 240
241 TLEHQTGIKR VVECHGSFSK CTCTRCGQKY DGNEIREEVL AMRVAHCKRC EGVIKPNIVF 300
301 FGEDLGREFH QHVTEDKHKV DLIVVIGSSL KVRPVALIPH CVDKNVPQIL INRESLPHYN 360
361 ADIELLGNCD DIIRDICFSL GGSFTELITS YDSIMEQQGK TKSQKPSQNK RQLISQEDFL 420
421 NICMKEKRND DSSDEPTLKK PRMSVADDSM DSEKNNFQEI QKHKSEDDDD TRNSDDILKK 480
481 IKHPRLLSIT EMLHDNKCVA ISAHQTVFPG AECSFDLETL KLVRDVHHET HCESSCGSSC 540
541 SSNADSEANQ LSRAQSLDDF VLSDEDRKNT IHLDLQRADS CDGDFQYELS ETIDPETFSH 600
601 LCEEMRI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
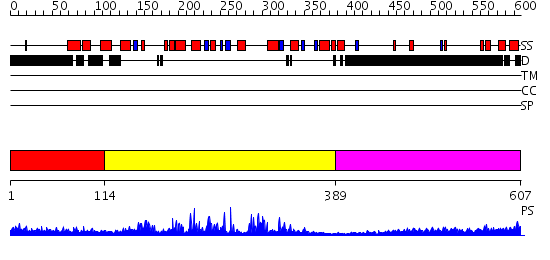
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..113] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [114..388] | 68.09691 | Sirt2 histone deacetylase | |
| 3 | View Details | [389..607] | 8.149996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)