













| General Information: |
|
| Name(s) found: |
acs-14 /
CE05585
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 544 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
propionate catabolic process, 2-methylcitrate cycle
[IEA teichoic acid biosynthetic process [IEA metabolic process [IEA menaquinone biosynthetic process [IEA siderophore biosynthetic process [IEA |
| Molecular Function: |
D-alanine-poly(phosphoribitol) ligase activity
[IEA AMP binding [IEA acetate-CoA ligase activity [IEA catalytic activity [IEA propionate-CoA ligase activity [IEA o-succinylbenzoate-CoA ligase activity [IEA (2,3-dihydroxybenzoyl)adenylate synthase activity [IEA ligase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPHDSTYPPV KISERPYHEM LLDAIDKTIA DGKNKIAFVS GDNPKHFITF EQLRKDAFAV 60
61 AMYLHSIGFK KDVAAVVLPN VWHYTSFFIG CAINGGAVSG ASALFTDYEL QRQFVDSRAK 120
121 VVFTYEDFLP KVLLAVKQSP NIQKIIVIPK PVGSSLPAGV VSWNEVVSTP VTALPQVPID 180
181 VHNDLLVLPY SSGTTGPPKG VMLSHYNFTS MISMYLAIDK SHILDVLDPN WDCYNEKALL 240
241 FLPFYHVYGF GLLNHCLLKG MTGIVMSHFE PNNFLTAVQN YKVRCLCLVP PIMVFLAKHP 300
301 ICDKFDLSSV QMIMAGAAPA GKDLIEELKR KYTNLKYIQQ GYGMTECSMA SHLPDLRNDQ 360
361 PYGSVGKLAS NLVMKIVEPG TDREQPVNQR GEICVRGPTI MLGYLGRPEA TASTVIDGWL 420
421 HTGDIGYLNE DGNLFIVDRL KELIKVKGLQ VPPAELEDLL LSHPKIRDCA VIGIPDAKAG 480
481 ELPKAFVVRA DNTLTEQEVK DFVKPKVSPY KQLEGGVEFI EEIPKSAAGK ILRRFLRDRS 540
541 SAKL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
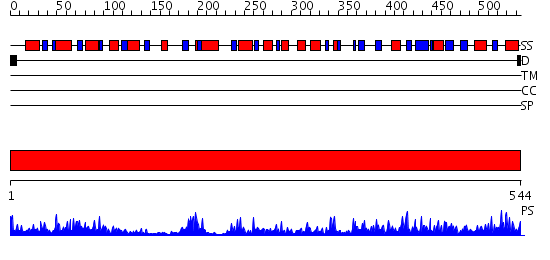
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..544] | 109.0 | FIREFLY LUCIFERASE IN COMPLEX WITH BROMOFORM |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.51 |
Source: Reynolds et al. (2008)