













| General Information: |
|
| Name(s) found: |
sir-2.2 /
CE02243
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 287 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromatin silencing complex
[IEA |
| Biological Process: |
DNA repair
[IMP regulation of transcription, DNA-dependent [IEA chromatin silencing [IEA |
| Molecular Function: |
DNA binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQKFVPEAA ELCENSLKKF ISLIGTVDKL LVISGAGIST ESGIPDYRSK DVGLYARIAH 60
61 KPIYFQDYMR SNRCRQRYWS RNFLAWPRFG QAAPNINHYA LSKWEASDRF QWLITQNVDG 120
121 LHLKAGSKMV TELHGSALQV KCTTCDYIES RQTYQDRLDY ANPGFKEEHV APGELAPDGD 180
181 IILPLGTEKG FQIPECPSCG GLMKTDVTFF GENVNMDKVN FCYEKVNECD GILSLGTSLA 240
241 VLSGFRFIHH ANMKKKPIFI VNIGPTRADH MATMKLDYKI SDVLKEM |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
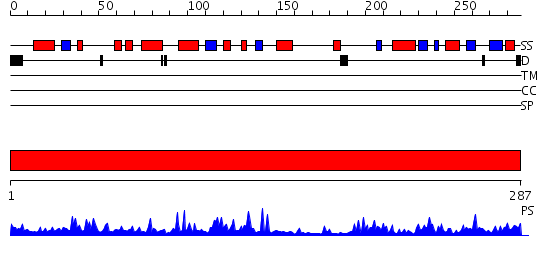
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..287] | 74.69897 | AF0112, Sir2 homolog (Sir2-AF2) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)