













| General Information: |
|
| Name(s) found: |
acs-7 /
CE00869
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 540 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
propionate catabolic process, 2-methylcitrate cycle
[IEA teichoic acid biosynthetic process [IEA metabolic process [IEA menaquinone biosynthetic process [IEA siderophore biosynthetic process [IEA |
| Molecular Function: |
D-alanine-poly(phosphoribitol) ligase activity
[IEA AMP binding [IEA acetate-CoA ligase activity [IEA catalytic activity [IEA propionate-CoA ligase activity [IEA o-succinylbenzoate-CoA ligase activity [IEA (2,3-dihydroxybenzoyl)adenylate synthase activity [IEA ligase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIFHGEQLEN HDKPVHEVLL ERFKVHQEKD PDNVAFVTAE NEDDSLGFQQ LGKKVLQISE 60
61 WFVENGYKKG DVFLLASYNN WRCFAAALGA WRAGLIVSAA ASQFTSFEMN YQIEDSQSQV 120
121 ILVDKHTLPV VQEACKNLKF VKQIISISAN PPSPVIKFDV LTSRLVRNLK MPLIDPKNDI 180
181 VFLPYSSGTT GKPKGVMISH LNFSMMLESS LRFFDANAKA IGLPPDFVLP YDLHFLPMYH 240
241 AMGMFRTLLT SYRGTTQIMF TKFDMELMLK NIEKYSIMVL SLVPAIAVRM LNSPLLQKYD 300
301 VSSLVSVTVG SAPFPESASK KLKQLLPNVN IVQGYGMTEL TFATHLQSPG SPDGSVGRLV 360
361 PGTSMKVKKE DGTLCGPHEI GELWIKGPQM MKGYWKKEQQ TNELLDEHGF MRTGDIVYFD 420
421 KNGETFICDR IKELIKVNAK QVAPAELESV ILEHDDVADV CVFGVDDASS GERPVACVVS 480
481 KRGRDMETSK AIMKHINQKL ARYKHIKEIE FVSEIMRTGT GKLLRRAMKK AFLDAKKAKL 540
541 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
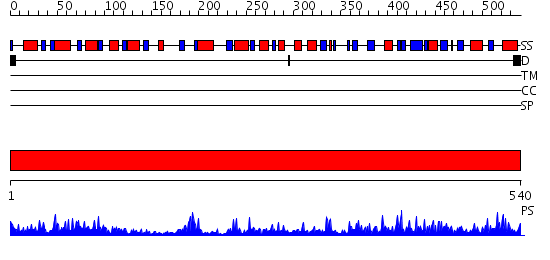
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..540] | 112.0 | FIREFLY LUCIFERASE IN COMPLEX WITH BROMOFORM |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)