













| General Information: |
|
| Name(s) found: |
SPBC1685.05
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 997 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
proteolysis
[ISS proteolysis involved in cellular protein catabolic process [NAS] |
| Molecular Function: |
serine-type endopeptidase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTPVSSDYRP NHIFLPKFRP DKITSECNTP QQQNVIAEGL YNVSEPFPIR NTDEERYLDT 60
61 KEIHHTWDNT IKNVVRSIVS IKGSALRSFD TESAGSFCAT GFVVNKTLGL ILSNRHVVSP 120
121 GPISARASFI NYEEIDIYPI YRDPVHDFGF FRYDPSSIRF HDVTEISLSP ESAKVGIDIR 180
181 IIGNDAGEKL SILSSTLARL DRPAPNYGID NYNDFNTFYY QAASGTSGGS SGSPVLDISG 240
241 AAVALNSGGS NSSASSFYLP LDRVVRALRC IENNTPITRG TLLTEFLHWS YDELSRIGLP 300
301 REFEYDCRTR VPSSTGLLVV SRVLRNSEVS KALEPGDILI AFKTDSHKST YIVDFVSLFE 360
361 VLDEMVGKTI ELHVYRPKRG FLTFQLTVQD LHNVTPSRFL EVGGAVLHDL SYQLARSYQF 420
421 SLNSGTYVAS SGMLNWSSGT RDFLVTRLAN KPTPTLDAFI DVLVQLTDNA RVPMHFRVLG 480
481 KYEEEFTIVT VDRHFFLASI FSRNDEKGTW DRQSLPPPQP SISRRPSVIP RPQEGSKSME 540
541 AIQNALVLVH CRMPYSINGF SSTKLYSGTG VIVSVVPPLI VVDRSVIPVD ICDIRLTFQS 600
601 MSAMGHLTFL DNRIAVVSCD YLPSNSVQLN FVADFLRTGD ECTLAALDED LQLLTKKTTV 660
661 RSVSVVETER SSPPRFRYVN CEVISLMDSL ASTGGLVFRE VGDDREIVAL WISVVHQDVG 720
721 GKDYTTKYGL SMSYILPVLE RLKLPPSARA QHVPTTAGVE WSHITLAGAS TLGLSQTRSS 780
781 EFYMKSRENG TIPRPLYVIS HLRPLLHKTS LGVGDILLEV NGKMITRLSD LHEFETESEI 840
841 KAVILRDGIE MEITIPLYPE YPTFSSRAIC WMGAIIHPTH SSVFEQVEPD VDLPGPEGVY 900
901 VGSILYGSPA LNMLRAAHWI VAVDGHDINT FDDFYHMLLE KPTDTFVQVK QMNRRGATSI 960
961 VSVRPDPLFW PTCIIERDSN GRWCTKHLQR KTKEVCS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
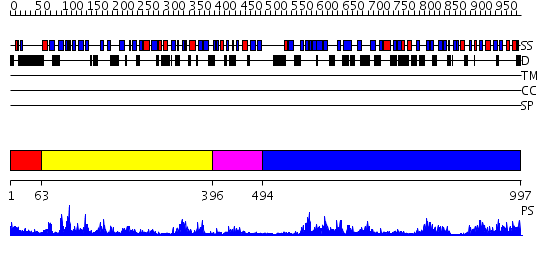
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..62] | 1.324996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [63..395] | 39.69897 | Crystal Structure of RV0983 from Mycobacterium tuberculosis- Proteolytically active form | |
| 3 | View Details | [396..493] | 6.0 | Structural Insights into the Inter-domain Chaperoning of Tandem PDZ Domains in Glutamate Receptor Interacting Proteins | |
| 4 | View Details | [494..997] | 73.522879 | Protease Do (DegP, HtrA), C-terminal domains; Protease Do (DegP, HtrA), catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)