













| General Information: |
|
| Name(s) found: |
fft1 /
SPAC20G8.08c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 944 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC nuclear chromatin [IC] chromatin remodeling complex [ISS |
| Biological Process: |
chromatin remodeling
[ISS |
| Molecular Function: |
ATP binding
[ISS DNA binding [IEA] ATP-dependent DNA helicase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRKDNKELI CIPSSSPPST PIREENYYLL TSSPIECSPI QQLDLSGSFK NYSTTSSRAN 60
61 GKKFGQLAMQ SRIFFDTTDH KKYVESSYAA YDPHDQPPER DVSLKESSNK INPSNFFDSE 120
121 QITKINAVKK RFPALDSEMI IATLEKFNWR ENITIHKLTF QKLLGRGNST INKSAQKLNN 180
181 QPIEKSSVDK ENAKRKRYVE EGTKQGQKKK PLRVIELSDE ETNEDDLLGQ SPTACTTDAN 240
241 IDNSIPENSD KIEEVSIESS GPSEVEDEMS EYDVRVLNFL NESTLQEIVE VSGCESKVVE 300
301 YFISKRPFPS LEKAEALCQR NAHAATGKRK KSDGRNVGRK LVNSTYEVLQ GFDAVDSLIA 360
361 KCERYGAMIS NTMRSWHNLF DDKKMEQFLN TSTGSISYEY NSQQPSSIAS GITLKSYQIV 420
421 GLNWLCLMYK AKLSGILADE MGLGKTCQVI SFLASLKEKG IQNRHLVVVP SSTLGNWLRE 480
481 FEKFCPSLRV ESYSGTQSER INKRYYLMDT DFDVLVTTYQ LASGSRDDRS FLRKQRFDIS 540
541 IFDEGHYLKN RMSERYKHLM NIPANFRLLI TGTPLQNNLK ELISLLAFML PKVFDNNMQG 600
601 LDIIYKIKTT SDGDIERAYL SQERISRAKT IMNPFILRRR KENVLSDLPP KIQHVEYCHM 660
661 EETQLSLYLS VLELKNLVNA NRENILMQLR KAALHQLLFR SQYNLETLSL MSKRILREDA 720
721 YLDANPQYIF EDMEVMSDFE LHKLADQYRH LHPFALKGKP WMDSAKVKKL CSLLKKSRPN 780
781 ERILIFSQFT QVLDILEYVL NTLDLEFLRL DGSTPVETRQ QLIDDFHTNE NYKVFLLSTK 840
841 SGGFGINLTC ANIVILFDCS FNPFDDMQAE DRAHRVGQTR PVHVYRLITK NTIEENIRRL 900
901 ANTKLTLESS LTTDSEKIQK EISGELMKSL QMDGRVDTMD GSVV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
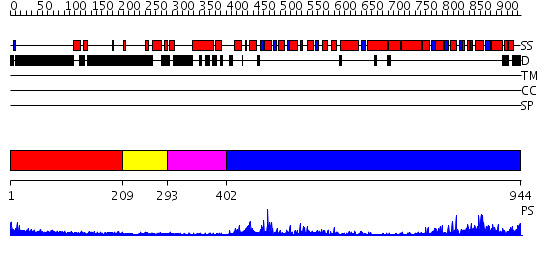
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..208] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [209..292] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [293..401] | 4.0 | Crystal structure of the UvsW helicase from Bacteriophage T4 | |
| 4 | View Details | [402..944] | 83.39794 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)