













| General Information: |
|
| Name(s) found: |
SPBC19C7.11
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 812 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloride channel complex
[IEA]
endosome [ISS membrane [IEA] integral to membrane [IEA] Golgi apparatus [ISS |
| Biological Process: |
transmembrane transport
[IEA]
cellular cation homeostasis [ISS chloride transport [IC |
| Molecular Function: |
chloride ion binding
[IEA]
voltage-gated chloride channel activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSQKRADSS SSFDQAERRS LDENERKKKY FFNREGSGID NFETDGMVFQ ASKAFDYYLL 60
61 TTFKDRVNEI INEQNEENVI DQSRWSKLWR IWNVGYSWFI LSIIGTTVGF AAYMLDIVTS 120
121 WLSDIRRGYC TSHWYYNEKF CCWYSETMGM FKHDLYNDLT FQGSSCTAWK PWTYKFSLNY 180
181 LIYTAFALLF VLCAAIMVRD VAPLAAGSGI SEIKCIISGF LRDSFLSFRV MLVKCVGLPL 240
241 AIASGLSVGK EGPSVHLATT IGHNISKIFK YAREGSIRYR DICVASAASG VAVAFGSPIG 300
301 GVLFGIEVGK FIEILTNSFK EMSGGYDPKM IVYSFFCCLS AVGVLHMLNP FRTGQVVLFE 360
361 VRYSGSWHFF ELLFFCFLGI FGGLYGEFVM RLFFLIQKLR KKYLSRWGVL DAAFVTVITS 420
421 LVSFLNPWLR LDMTLGMELL FQECKSSSSP ELINLCDPSL RTKNTILLLI ATFARTIFVT 480
481 FSYGAKVPAG IFVPSMAVGA SFGYMIGLIA EMIYQRFPNS VLFLACHGSE SCITPGTYAL 540
541 LGAAASLSGI MHLTVTIVVI MFELTGALNF ILPTVLVVAL ANSIGNMLGK TGIADRSIEI 600
601 NGLPLLEPEK SINSSNTINI PITEVMASNL ITIPSIGFTW RKLLGMMEGY DFSGYPVVLD 660
661 SRSNYLIGYL KKSSLKSSFE AAKLEPSFTF DQQLCFGKVD SVGDSKSSKF GESDRIDLSA 720
721 YMDVNPISVL HTQSIANVAV LFEVLSPSVI FIEKDGNLVG LISKKDLLLA SKYYQEDNDL 780
781 VNPANRHRAV STERINDSYE NIELKPLKLN IE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
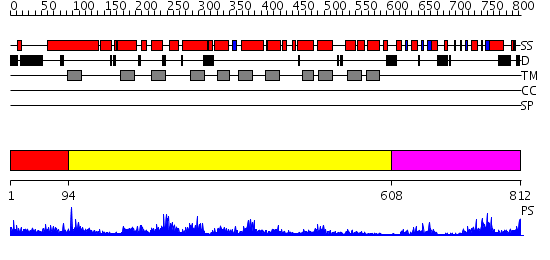
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | 2.16499 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [94..607] | 56.522879 | Clc chloride channel | |
| 3 | View Details | [608..812] | 26.69897 | Crystal structure of Inosine-5'-monophosphate dehydrogenase (TM1347) from THERMOTOGA MARITIMA at 2.18 A resolution |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|