













| General Information: |
|
| Name(s) found: |
SPAC4G9.15
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 341 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to membrane
[IEA]
endoplasmic reticulum [IDA |
| Biological Process: |
fatty acid elongation
[ISS |
| Molecular Function: |
ketoreductase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGEVLANKS CCGAVVTAFS VIGIVFTILK FTSFASFVYK TFFAKGVKLS VYGAKKGYWA 60
61 VVTGATDGIG KEYATQLAMS GFNVVLISRT QEKLDALAKE LETVAKVKTR TIAIDYTKTT 120
121 AETFEKLHQD LVGTPITVLI NNVGQSHYMP TSFAETTVKE MDDIMHINCF GTLHTTKAVL 180
181 SIMLRERQKN EKGPRCLILT MGSFAGLLPS PYLSTYAGSK AFLSNWSASL GEEVKKQGID 240
241 VWCFNSYLVV SAMSKVRRPT LTIPTPKKFV RAALSSIGLQ RGGTNPYISQ PYPSHAVMSW 300
301 SLEQLLGSAK GFVVSQVAAM HLSIRKRALR KEARLQAQNQ A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
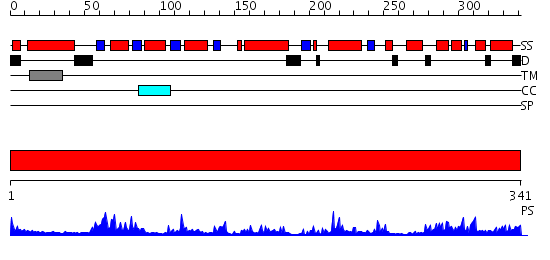
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..341] | 52.522879 | Crystal structure of peroxisomal trans 2-enoyl CoA reductase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|