













| General Information: |
|
| Name(s) found: |
brc1 /
SPBC582.05c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 878 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
DNA repair
[ISS chromosome organization [IGI regulation of DNA recombination [NAS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKIKLLNVK TPNHYTIIFK VVAYYSALQP NQNELRKKEL FIKNDGKALS FPYDWKLATH 60
61 VICDDFSSPN VQEGSKRSLR LAKTNWIRDC VDKNTLLNYS FYSCNPYLLF KGICASSCQI 120
121 DSYQSSLIDD ALETFGGRFS KGLMKSMTHL FTYSGMGAKC KKVLDKPSLS IKLIHPQWLL 180
181 DCLQFGQLID QDPYLFPNPS YKKNDSSISK AEPTSLFRNV LHGKRIYFSN DLNLPTNFRH 240
241 SLQKFSVGIG AKIAESINDC DIFIGLKRDT IEFNLASNKN TTIGTISWLL NLFVLGSWKS 300
301 PLLNALHYPF PSVGFLKDQM VAVTNYTDAA RIYLEKLLLA CGATYTKDLK PTNTLLIAAS 360
361 SYGQKYGAAK VWNIPTVHHS WLYSSFKNLS SQAFTDFPVP LDDSYMDFIF PCPLNVEKGS 420
421 FEDTLKSSLT KGNSEVLLDD LSDPSVSSIK GNKTNEELEK EFKSTSDNFG KHIILTSSFS 480
481 NQSADKGSSL AAEDDRNDEG STITGVNREL QDEGRLEIDA KSSKTNTPPS PLLVGTPSKE 540
541 SLKEASSDDE LPVLATKLVD NVIKEKSPLS LTPKVVVPSH KETYTDEKKL IDELDRVNPL 600
601 NSSQLLRSKR KSAATALSML QNVIMPDVLA FEREKKRRQT HRSVSSGEVS RESSESRNTN 660
661 AKASKRVYIT FTGYDKKPSI DNLKKLDMSI TSNPSKCTHL IAPRILRTSK FLCSIPYGPC 720
721 VVTMDWINSC LKTHEIVDEE PYLLNDPEKE LELGCTLESA LKRARAQGPS LLEDYVVYLT 780
781 SKTVAPENVP AVISIVKSNG GVCSTLNVYN KRLARHLEDG NVVLITCNED SHIWTNFLDN 840
841 ASQNKTIFLQ NYDWLIKTVL RQEIDVNDRI ADEFARAV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
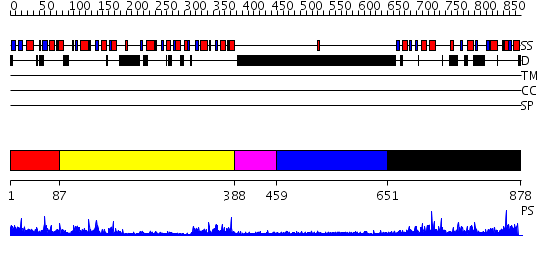
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | 21.39794 | The third BRCA1 C-terminus (BRCT) domain of Similar to S.pombe rad4+/cut5+ product | |
| 2 | View Details | [87..387] | 35.39794 | No description for 2owoA was found. | |
| 3 | View Details | [388..458] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [459..650] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [651..878] | 41.0 | Crystal structure of the MDC1 brct repeat in complex with the histone tail of gamma-H2AX |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)