













| General Information: |
|
| Name(s) found: |
srs2 /
SPAC4H3.05
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 887 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA spindle pole body [IDA spindle [IDA |
| Biological Process: |
recombinational repair
[IGI cellular response to UV [IMP replication fork processing [TAS postreplication repair [IGI |
| Molecular Function: |
DNA binding
[IEA]
ATP binding [IEA] ATP-dependent DNA helicase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METKSSYLKF LNEEQRISVQ SPHKYTQILA GPGSGKTRVL TARVAYLLQK NHIAAEDLII 60
61 ATFTNKAANE IKLRIEAILG NSEASKLISG TFHSIAYKYL VKYGKHIGLS SNWLIADRND 120
121 TQAIMKRLLD SLKKAKNPIA SGIRGQELTP QNALNRITKL KSNGLLVKPG MDQLSLINGL 180
181 EEPPKELQSH QSVELYRLYQ TSLWKNNLAD FDDLLLNFIL LLQKQPDCVR NIKHILIDEF 240
241 QDTSKIQYFL VKLLALQNSD ITIVGDPDQS IYGFRSAEIR NLNQMSEDFE GTQVLHLERN 300
301 YRSAKPILEL ALSIISQDKS RPKKGLKSNH ISSLKPHYRL FETNNKESYW IAREIKRIVG 360
361 SCPELIFYND IAILVRSSSL TRSLEHALSE LGVPYRMVGV NKFFDREEIR DLIAYLRVLA 420
421 NKDSTSLIRV INVPPRNIGK TKIDRIIFES ERRGLTFWQT LNEVKNENIL LSQRNDKSFL 480
481 KSLKSFLCSI SKLENRYLSN GHSATLSDLL LGILSEIKYY EYLVRKNKET VEEKWENVME 540
541 LVQQSDNISC IFYELDYKIS TIVLLQNFLT QIALVNEEQK EGESQKVTIS TLHAAKGLEW 600
601 PVVFLPCLCE NIIPHSRSDD LDEERRLLYV GATRAQALLY LSSFKSVTGM FADMQNSDNV 660
661 QDVSPFLKGE EMKRWVMESE IVFNEKIASE IGTILGRKSY GKITNLSGIG SNANHNGTKF 720
721 ENLGFQCCRV LAEAELKKRE RVKSVNDYNK DETNFRKHNA KRSKTDIRSW FEKKQPIDSD 780
781 VEISEPSRSA SIMVANKDLN DRSFETVNRI VSTRASTTNA SFMSSVRQNL GRGPSTKDQV 840
841 INRTLREGHQ DVVQHTDLNQ SNTKVASARP AGSRKRLGVR LRVSRML |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
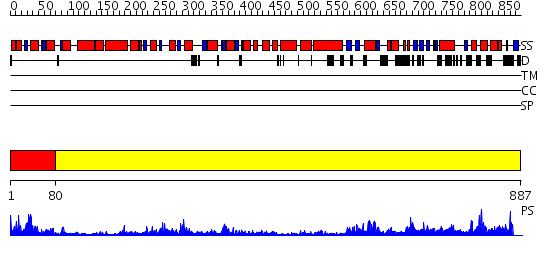
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] | 145.0 | STRUCTURE OF DNA HELICASE | |
| 2 | View Details | [80..887] | 42.0 | RecBCD:DNA complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)