













| General Information: |
|
| Name(s) found: |
SPAC12G12.16c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 496 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
DNA repair
[ISS |
| Molecular Function: |
magnesium ion binding
[IEA]
DNA binding [IEA] endonuclease activity [IEA] nuclease activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGVHGLLPII RKVASEGIRW LQPEFFSKYY PVLAVDGNLL LYRFFRSPNT PLHTNYQHVL 60
61 WALKLSRYCR KLKTSPIVVF DNLKPNDFKA EEHEKRSLIS SQIVNQRQVV QEQMYFLCNL 120
121 KNCLIDNNFP TGNFYYEIGP LLSQIGKLLF DLRSLNERDN PFHAQNNAEN LENATFVRLI 180
181 VDKLNDREKT LMIQEKKNHL IHSLRQLLAF SEINDFPSEI RSYLEFILSN LDLECLTLCL 240
241 KIIKGILTLD ELQKAIKLKQ TELDKLERRL YRPSPQNIFE IFEILKILGI PASFSPIGVE 300
301 AEAFASAISQ NNLAYAVATQ DTDVLLLGSS MISNFLDLND NFHLPLQIMD PRKIAQELNL 360
361 TFDGFQDYCL MCGTDFTSRI PKIGPVRALK LIRYYGNAFD VLKALNVEEK YIIPTDYIKK 420
421 FLTAKKLFTD LPSNNELFSF IHNIPKEFLH LSDSTYLDLE KQVLRIFNVQ IEELDAKTPW 480
481 YYHYELEKDV KSTYEY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
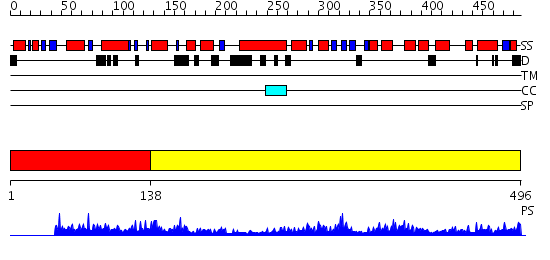
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..137] | 3.35 | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | |
| 2 | View Details | [138..496] | 75.0 | Crystal structure of the human FEN1-PCNA complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)