













| General Information: |
|
| Name(s) found: |
SPAC16C9.04c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 489 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA CCR4-NOT core complex [RCA CCR4-NOT complex [ISS |
| Biological Process: |
negative regulation of transcription
[IC mRNA catabolic process [IC nuclear-transcribed mRNA poly(A) tail shortening [ISS RNA elongation from RNA polymerase II promoter [ISS] protein ubiquitination [ISS regulation of transcription from RNA polymerase II promoter [ISS |
| Molecular Function: |
ubiquitin-protein ligase activity
[ISS zinc ion binding [IEA] RNA binding [ISS transcription repressor activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLKTQEIYSS EDEDDMCCPL CMEEIDISDK NFKPCQCGYR VCRFCWHHIK EDLNGRCPAC 60
61 RRLYTEENVQ WRPVTAEEWK MDLHRKNERK KREKERKEVE LSNRKHLANI RVVQKNLAYV 120
121 NGLSPKVANE ENINVLKGPE YFGQYGKIIK IAINKKAAAN SANGHVGVYI TYQRKEDAAR 180
181 AIAAIDGSVS DGRHLRASYG TTKYCTSYLR NQQCPNPSCM YLHEPGDEVD SYTKEDLASL 240
241 QHTRPLSTKP NVVNGATHSP SPSLPFKTPL LPVTKTPLEE ANSSPAAQNQ HITTVDHVHP 300
301 QVSMTPSLST NNTATSVPAP YSSAASVNVV PGHATTILHH EESSALPPTA AWAKLSPSVL 360
361 QERLRAAVNQ QPLDALKSSS TQTSIPKIQK LKAAKLPSEE ENTTKWLNKA INDLVSSLSK 420
421 INFSTEGTEF DKKQIEMIQN LPPLFVFNAR SVIDKEVVPE QEKSAENQPP TSLGINNGNP 480
481 VMPPPGFQS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
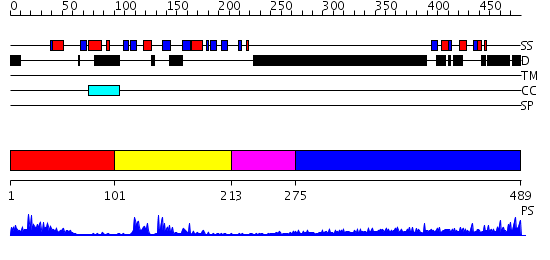
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..100] | 11.69897 | Not-4 N-terminal RING finger domain | |
| 2 | View Details | [101..212] | 5.522879 | Solution structure of the RNA recognition motif of CNOT4 | |
| 3 | View Details | [213..274] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [275..489] | 3.522879 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)