













| General Information: |
|
| Name(s) found: |
smc5 /
SPAC14C4.02c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1065 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cell division site [IDA Smc5-Smc6 complex [IDA spindle pole body [IDA spindle [IDA cytosol [IDA |
| Biological Process: |
replication fork protection
[IC double-strand break repair via homologous recombination [TAS |
| Molecular Function: |
ATP binding
[ISS protein binding [IPI protein heterodimerization activity [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MILTRESYAL GSIVRIKLVN FVTYDYCELF PGPYLNLIIG PNGTGKSTIV SAICIGLGWP 60
61 PKLLGRAKEA REFIKYGKNT ATIEIEMKYR DDETVTITRQ ISQDKSSSFS INREACATSS 120
121 ITSLMDTFNV QLNNLCHFLP QDRVAEFAQL DPYSRLMETE RAIDHEGLLP AHEKLIDLRK 180
181 REREILQNKN QGQSTLNSLK DRQQALEKEV NIFKEREKIK SYIEMLGLAK MLVIYREKTN 240
241 VFNQLRADKK KLKKDLKDLV EEFQPILDKG EELRSDLKLK DDTFNDYSSA SMELNTSNLR 300
301 ARASFSNFME NEKKLYEKVN TNRTLLRNAN LTLNEAQQSV KSLTERQGPR PSDNGVQDLQ 360
361 EKMQEVNAEK LQHENEKLES SHELGSIRTL KAQKLIDLDN IKRELSYYND ATKRKLDFMS 420
421 SAPGWEDAYQ TYQLLKEYES AFEAPAYGPI YMNLKCKEKG FAALIEGFFR TDTFRTFIMS 480
481 NYNDYLKLMD LITSKTKYTP TIREFSSERK KKIEDFEPPC SREKLQSFGF DGYVIDFLEG 540
541 PEVVLVALCH MLKIHQIPIA KRELPPASVN ALNNFRLANG DPVLKTYLAG SSIHLVFRSA 600
601 YGDREITRRT DPLPSRSIYF SENVEMDLVK RKEEQLNAQL SQLENLQNEE RKLQEKVNEH 660
661 ESLLSRTNDI LSTLRKERDE KLIPIHEWQQ LQERIEHQTL LLRQREKVPE QFAAEIEKNE 720
721 DIRKENFEAL MNSVLKVKEN SIKATNNFEK MLGSRLNVIE AKYKLEKHEM DANQVNARLT 780
781 EVQDRLKDIT DKLASAREDA MSLYGSVVDS LQTQSSDRQT AITELNEEFA TSSEVDNKIS 840
841 IEETKLKFMN VNSYVMEQYD ARKKEIEELE SKMSDFDQSV EELQDEMNSI KEDWVSKLEE 900
901 NVQCISDRFS KGMSGMGYAG EVRLGKSDDY DKWYIDILVQ FREEEGLQKL TGQRQSGGER 960
961 SVSTIMYLLS LQGLAIAPFR IVDEINQGMD PRNERVVHRH IVNSVCDNAV SQYFLVTPKL1020
1021 LPDLTYHRNL KVLCICNGAW LPATFRTSLS TYFEKLKKSA LISSS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
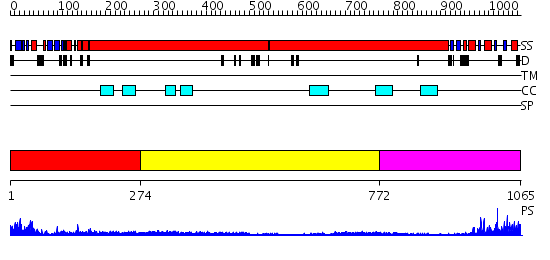
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..273] | 21.0 | Sc Smc1hd:Scc1-C complex, ATPgS | |
| 2 | View Details | [274..771] | 1.69 | Tropomyosin | |
| 3 | View Details | [772..1065] | 4.045757 | No description for 2iw3A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)