













| General Information: |
|
| Name(s) found: |
pot1 /
SPAC26H5.06
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 555 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA telosome [IC cytosol [IDA |
| Biological Process: |
chromosome segregation
[IMP telomere maintenance via telomerase [IMP telomere capping [IDA |
| Molecular Function: |
protein binding
[IPI single-stranded telomeric DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGEDVIDSLQ LNELLNAGEY KIGELTFQSI RSSQELQKKN TIVNLFGIVK DFTPSRQSLH 60
61 GTKDWVTTVY LWDPTCDTSS IGLQIHLFSK QGNDLPVIKQ VGQPLLLHQI TLRSYRDRTQ 120
121 GLSKDQFRYA LWPDFSSNSK DTLCPQPMPR LMKTGDKEEQ FALLLNKIWD EQTNKHKNGE 180
181 LLSTSSARQN QTGLSYPSVS FSLLSQITPH QRCSFYAQVI KTWYSDKNFT LYVTDYTENE 240
241 LFFPMSPYTS SSRWRGPFGR FSIRCILWDE HDFYCRNYIK EGDYVVMKNV RTKIDHLGYL 300
301 ECILHGDSAK RYNMSIEKVD SEEPELNEIK SRKRLYVQNC QNGIEAVIEK LSQSQQSENP 360
361 FIAHELKQTS VNEITAHVIN EPASLKLTTI STILHAPLQN LLKPRKHRLR VQVVDFWPKS 420
421 LTQFAVLSQP PSSYVWMFAL LVRDVSNVTL PVIFFDSDAA ELINSSKIQP CNLADHPQMT 480
481 LQLKERLFLI WGNLEERIQH HISKGESPTL AAEDVETPWF DIYVKEYIPV IGNTKDHQSL 540
541 TFLQKRWRGF GTKIV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
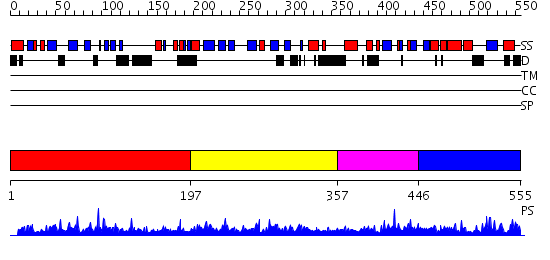
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..196] | 56.0 | Crystal structure of Pot1 (protection of telomere)- ssDNA complex | |
| 2 | View Details | [197..356] | 3.15 | Crystal structure of human POT1 bound to telomeric single-stranded DNA (TTAGGGTTAG) | |
| 3 | View Details | [357..445] | 2.03399 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [446..555] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)