













| General Information: |
|
| Name(s) found: |
pabp /
SPAC57A7.04c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 653 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA cytosol [IDA |
| Biological Process: |
mRNA export from nucleus
[IMP regulation of translational initiation [ISS] |
| Molecular Function: |
poly(A) RNA binding
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPSTDLKKQA DAAVESDVNT NNEAVESSTK EESSNTPSTE TQPEKKAEEP EAAAEPSEST 60
61 STPTNASSVA TPSGTAPTSA SLYVGELDPS VTEAMLFELF NSIGPVASIR VCRDAVTRRS 120
121 LGYAYVNFHN MEDGEKALDE LNYTLIKGRP CRIMWSQRDP SLRKMGTGNV FIKNLDPAID 180
181 NKALHDTFSA FGKILSCKVA VDELGNAKGY GFVHFDSVES ANAAIEHVNG MLLNDKKVYV 240
241 GHHVSRRERQ SKVEALKANF TNVYIKNLDT EITEQEFSDL FGQFGEITSL SLVKDQNDKP 300
301 RGFGFVNYAN HECAQKAVDE LNDKEYKGKK LYVGRAQKKH EREEELRKRY EQMKLEKMNK 360
361 YQGVNLFIKN LQDEVDDERL KAEFSAFGTI TSAKIMTDEQ GKSKGFGFVC YTTPEEANKA 420
421 VTEMNQRMLA GKPLYVALAQ RKEVRRSQLE AQIQARNQFR LQQQVAAAAG IPAVQYGATG 480
481 PLIYGPGGYP IPAAVNGRGM PMVPGHNGPM PMYPGMPTQF PAGGPAPGYP GMNARGPVPA 540
541 QGRPMMMPGS VPSAGPAEAE AVPAVPGMPE RFTAADLAAV PEESRKQVLG ELLYPKVFVR 600
601 EEKLSGKITG MLLEMPNSEL LELLEDDSAL NERVNEAIGV LQEFVDQEPG FTE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
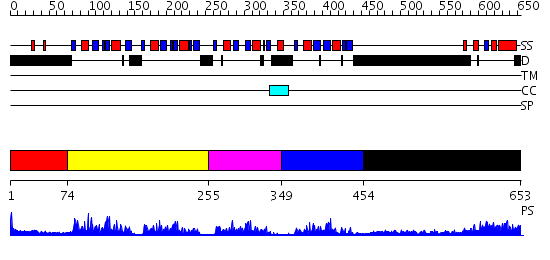
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..73] | 18.0 | CG8781 protein | |
| 2 | View Details | [74..254] | 50.221849 | Poly(A)-binding protein | |
| 3 | View Details | [255..348] | 6.69897 | Splicing factor U2AF 65 KDa subunit | |
| 4 | View Details | [349..453] | 4.63 | No description for 2f3jA was found. | |
| 5 | View Details | [454..653] | 32.522879 | poly(A) binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)