













| General Information: |
|
| Name(s) found: |
myh1 /
SPAC26A3.02
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 461 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
DNA damage checkpoint
[IGI DNA repair [IC base-excision repair [IC cellular response to UV [IMP |
| Molecular Function: |
DNA binding
[IDA protein binding [IPI iron ion binding [IEA] endonuclease activity [IEA] oxidized base lesion DNA N-glycosylase activity [TAS purine-specific mismatch base pair DNA N-glycosylase activity [IMP DNA N-glycosylase activity [IDA 4 iron, 4 sulfur cluster binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDSNHSLDL HSYTQLEVER FRESLIQFYD KTKRILPWRK KECIPPSEDS PLEDWEQPVQ 60
61 RLYEVLVSEI MLQQTRVETV KRYYTKWMET LPTLKSCAEA EYNTQVMPLW SGMGFYTRCK 120
121 RLHQACQHLA KLHPSEIPRT GDEWAKGIPG VGPYTAGAVL SIAWKQPTGI VDGNVIRVLS 180
181 RALAIHSDCS KGKANALIWK LANELVDPVR PGDFNQALME LGAITCTPQS PRCSVCPISE 240
241 ICKAYQEQNV IRDGNTIKYD IEDVPCNICI TDIPSKEDLQ NWVVARYPVH PAKTKQREER 300
301 ALVVIFQKTD PSTKEKFFLI RKRPSAGLLA GLWDFPTIEF GQESWPKDMD AEFQKSIAQW 360
361 ISNDSRSLIK KYQSRGRYLH IFSHIRKTSH VFYAIASPDI VTNEDFFWIS QSDLEHVGMC 420
421 ELGLKNYRAA LEIKKRKVTS LSNFKEPKLT SARRIVTKAE C |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
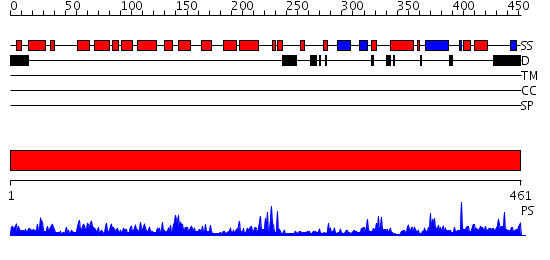
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..461] | 75.69897 | MutY adenine glycosylase in complex with DNA containing an abasic site |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)