













| General Information: |
|
| Name(s) found: |
mcm7 /
SPBC25D12.03c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 760 amino acids |
Gene Ontology: |
|
| Cellular Component: |
replication fork
[RCA pre-replicative complex [IC DNA replication preinitiation complex [IC] MCM complex [IDA |
| Biological Process: |
DNA replication initiation
[IDA S phase of mitotic cell cycle [IC DNA strand elongation involved in DNA replication [TAS |
| Molecular Function: |
DNA binding
[IEA]
ATP binding [IEA] ATP-dependent DNA helicase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALPTIDLKI PEYEECQHKI TDFLSHFKQE QVQDGQQNQD ISMSDAGDEP FLKSKYMDIL 60
61 QKISNRESNV INVDLNDLYE FDPSDTQLLH NIESNAKRFV ELFSQCADAL MPPPTVEINY 120
121 RNEVLDVIMQ QRVQRNENID PEHKGFPPEL TRGYDLYFRP VTRNKKPFSV RDLRGENLGS 180
181 LLTVRGIVTR TSDVKPSLTV NAYTCDRCGY EVFQEIRQKT FLPMSECPSD ECKKNDAKGQ 240
241 LFMSTRASKF LPFQEVKIQE LTNQVPIGHI PRSLTVHLYG AITRSVNPGD IVDISGIFLP 300
301 TPYTGFRAMR AGLLTDTYLE CHYVSQIIKN YTNIEKTPQS EAAIAELNQG GNVYEKLAKS 360
361 IAPEIYGHED VKKALLLLLV GGVTKELGDG MRIRGDINIC LTGDPGVAKS QLLKYISKVA 420
421 PRGVYTTGRG SSGVGLTAAV MRDPVTDEMV LEGGALVLAD NGICCIDEFD KMDESDRTAI 480
481 HEVMEQQTIS ISKAGITTTL NARTSILAAA NPLYGRYNPK VAPIHNINLP AALLSRFDIL 540
541 FLILDTPSRE TDEHLAQHVT YVHMHNEQPK MDFEPLDPNM IRHYISSARQ YRPVVPKDVC 600
601 DYVTGAYVQL RQNQKRDEAN ERQFAHTTPR TLLAILRMGQ ALARLRFSNR VEIGDVDEAL 660
661 RLMSVSKSSL YDDLDPSSHD TTITSKIYKI IRDMLNSIPD VEGNERSLTL RAIRERVLAK 720
721 GFTEDHLINT IQEYTDLGVL LTTNNGQTIM FLDPDLHMEN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
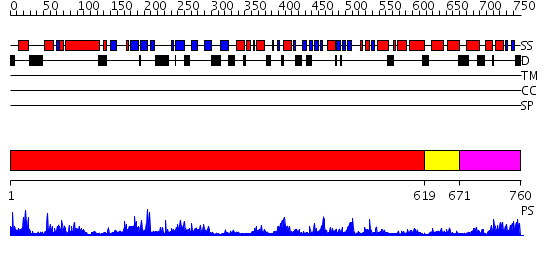
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..618] | 38.69897 | N-terminal, ClpS-binding domain of ClpA, an Hsp100 chaperone; ClpA, an Hsp100 chaperone, AAA+ modules | |
| 2 | View Details | [619..670] | 22.69897 | No description for 2qp9X was found. | |
| 3 | View Details | [671..760] | 50.69897 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)