













| General Information: |
|
| Name(s) found: |
mcm6 /
SPBC211.04c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 892 amino acids |
Gene Ontology: |
|
| Cellular Component: |
replication fork
[RCA pre-replicative complex [IC nuclear chromatin [IDA DNA replication preinitiation complex [IC] cytosol [IDA MCM complex [IDA |
| Biological Process: |
DNA replication initiation
[IDA S phase of mitotic cell cycle [IC DNA strand elongation involved in DNA replication [TAS |
| Molecular Function: |
DNA binding
[IEA]
ATP binding [IEA] ATP-dependent DNA helicase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSLASQGNN ASTPAYRYGF QTSEVGDRPT VSMPSQPESS AMLEDDGMVK RKPFAAIGEA 60
61 IPKVIDTTGE SVREAFEEFL LSFSDDRVAG GDALPSASQE KYYVQQIHGL AMYEIHTVYV 120
121 DYKHLTSYND VLALAIVEQY YRFSPFLLRA LQKLIEKFEP EYYRSSLSRE NASLSPNFKA 180
181 SDKTFALAFY NLPFRSTIRD LRTDRIGRLT TITGTVTRTS EVRPELAQGT FICEECHTVV 240
241 SNVEQAFRYT EPTQCPNELC ANKRSWRLNI SQSSFQDWQK VRIQENSNEI PTGSMPRTLD 300
301 VILRGDIVER AKAGDKCAFT GILIAVPDVS QLGIPGVKPE AYRDSRNFGG RDADGVTGLK 360
361 SLGVRDLTYK LSFLACMVQP DDANDKSGAD VRGDGSQGIE EQDEFLQSLS QEEIDDLRAM 420
421 VHSDHIYSRL TNSLAPSVYG HEIIKKGILL QLMGGVHKLT PEGINLRGDL NICIVGDPST 480
481 SKSQFLKYVC NFLPRAIYTS GKASSAAGLT AAVVKDEETG DFTIEAGALM LADNGICAID 540
541 EFDKMDLSDQ VAIHEAMEQQ TISIAKAGIQ ATLNARTSIL AAANPIGGRY NRKTTLRNNI 600
601 NMSAPIMSRF DLFFVVLDEC NESVDRHLAK HIVDIHRLRD DAMQPEFSTE QLQRYIRYAR 660
661 TFKPKLNTES CAEIVKKYKQ LRMDDAQGAG KNSYRITVRQ LESMIRLSEA IARANCVDDI 720
721 TPAFVNEAYS LLRQSIIHVE RDDIEVEEDD AEAQELENDN TNTTNGNDNV SSEEALQKPK 780
781 VKITYDKYVS IMNGILQVLR QRSTEGVDGV PAGDLVQSYL ELREDQFHTE EDIIYEVGLV 840
841 RKVLTRLVHE SIIMEIQNLT DSAVRLPFEE RVFSIHPNCD IDALLSNGDV PN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
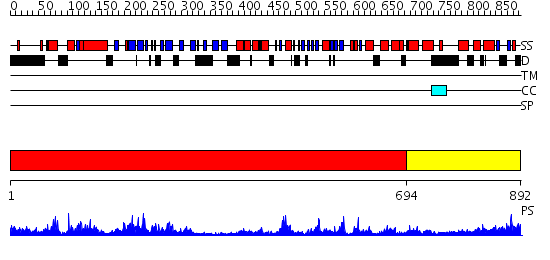
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..693] | 61.09691 | Crystal Structure Analysis of ClpB | |
| 2 | View Details | [694..892] | 46.09691 | EDTA treated |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)