













| General Information: |
|
| Name(s) found: |
tuf1 /
SPBC9B6.04c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 439 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[ISS mitochondrion [IDA |
| Biological Process: |
mitochondrial translation
[RCA translational elongation [IGI positive regulation of translational elongation [IC mitochondrial genome maintenance [IMP |
| Molecular Function: |
GTP binding
[ISS translation elongation factor activity [ISS GTPase activity [ISS tRNA binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSAKATSLL FQGFRKNCLR LNRISFASGL INRFTVPART YADEKVFVRK KPHVNIGTIG 60
61 HVDHGKTTLT AAITKCLSDL GQASFMDYSQ IDKAPEEKAR GITISSAHVE YETANRHYAH 120
121 VDCPGHADYI KNMITGAATM DGAIIVVSAT DGQMPQTREH LLLARQVGVK QIVVYINKVD 180
181 MVEPDMIELV EMEMRELLSE YGFDGDNTPI VSGSALCALE GREPEIGLNS ITKLMEAVDS 240
241 YITLPERKTD VPFLMAIEDV FSISGRGTVV TGRVERGTLK KGAEIEIVGY GSHLKTTVTG 300
301 IEMFKKQLDA AVAGDNCGLL LRSIKREQLK RGMIVAQPGT VAPHQKFKAS FYILTKEEGG 360
361 RRTGFVDKYR PQLYSRTSDV TVELTHPDPN DSDKMVMPGD NVEMICTLIH PIVIEKGQRF 420
421 TVREGGSTVG TALVTELLD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
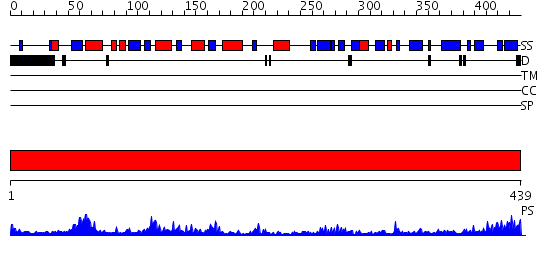
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..439] | 102.0 | Elongation factor Tu (EF-Tu), domain 2; Elongation factor Tu (EF-Tu); Elongation factor Tu (EF-Tu), N-terminal (G) domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)