













| General Information: |
|
| Name(s) found: |
dld1 /
SPAC1002.09c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 511 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial pyruvate dehydrogenase complex
[ISS mitochondrial oxoglutarate dehydrogenase complex [IC mitochondrion [IDA glycine cleavage complex [ISS mitochondrial nucleoid [ISS |
| Biological Process: |
acetyl-CoA biosynthetic process from pyruvate
[ISS leucine catabolic process [ISS valine catabolic process [ISS L-serine biosynthetic process [ISS glycine decarboxylation via glycine cleavage system [ISS isoleucine catabolic process [ISS 2-oxoglutarate metabolic process [IC |
| Molecular Function: |
disulfide oxidoreductase activity
[ISS FAD binding [IEA] oxoglutarate dehydrogenase (succinyl-transferring) activity [ISS pyruvate dehydrogenase (acetyl-transferring) activity [ISS glycine dehydrogenase (decarboxylating) activity [ISS dihydrolipoyl dehydrogenase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLNSVIKRSA LCRFKFTCLQ VSECRPAQIE ISKRLYSAKA SGNGEYDLCV IGGGPGGYVA 60
61 AIRGAQLGLK TICVEKRGTL GGTCLNVGCI PSKALLNNSH IYHTVKHDTK RRGIDVSGVS 120
121 VNLSQMMKAK DDSVKSLTSG IEYLFKKNKV EYAKGTGSFI DPQTLSVKGI DGAADQTIKA 180
181 KNFIIATGSE VKPFPGVTID EKKIVSSTGA LSLSEVPKKM TVLGGGIIGL EMGSVWSRLG 240
241 AEVTVVEFLP AVGGPMDADI SKALSRIISK QGIKFKTSTK LLSAKVNGDS VEVEIENMKN 300
301 NKRETYQTDV LLVAIGRVPY TEGLGLDKLG ISMDKSNRVI MDSEYRTNIP HIRVIGDATL 360
361 GPMLAHKAED EGIAAVEYIA KGQGHVNYNC IPAVMYTHPE VAWVGITEQK AKESGIKYRI 420
421 GTFPFSANSR AKTNMDADGL VKVIVDAETD RLLGVHMIGP MAGELIGEAT LALEYGASAE 480
481 DVARVCHAHP TLSEATKEAM MAAWCGKSIH F |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
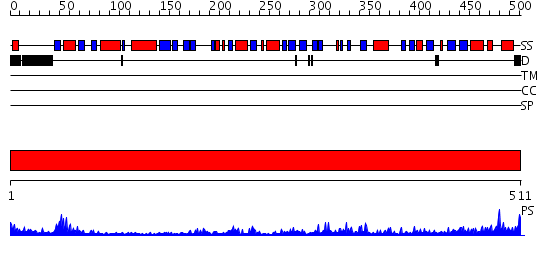
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..511] | 102.0 | Dihydrolipoamide dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)