













| General Information: |
|
| Name(s) found: |
cul3 /
SPAC24H6.03
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 785 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA SCF ubiquitin ligase complex [ISS |
| Biological Process: |
protein ubiquitination during ubiquitin-dependent protein catabolic process
[IC regulation of mitotic cell cycle [IC] |
| Molecular Function: |
ubiquitin protein ligase binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQRSAKLKIR APRKFSANQV DFATHWEVLQ RAIGDIFQKS TSQLSFEELY RNAYILVLHK 60
61 YGEKLYNHVQ DVIRSRLKEE TVPAIYKNYD ASLLGNALLD IRKNDSYSTS WSRSLEAAHR 120
121 FLSSLVNSWK DHIVSMQMIS SVLKYLDKVY SKSADKVPVN ENGIYIFREV VLLNSFEIGE 180
181 KCVETILILV YLERKGNTIN RPLINDCLDM LNSLPSENKK ETLYDVLFAP KFLSYTRNFY 240
241 EIESSTVIGV FGVVEYLKKA EKRFEEEKER SKNYLFTKIA SPLLSVVEDE LLSKHLDDLL 300
301 ENQSTGFFSM IDSSNFEGLQ LVYESFSRVE LGVKSLKKYL AKYVAHHGKL INETTSQALE 360
361 GKMAVGRLSS NATMATLWVQ KVLALWDRLN TIISTTMDAD RSILNSLSDA FVTFVDGYTR 420
421 APEYISLFID DNLKKDARKA IEGSIEATLQ NSVTLFRFIS EKDVFEKYYK THLAKRLLNN 480
481 RSISSDAELG MISRLKQEAG NVFTQKLEGM FNDMNLSQEL LQEYKHNSAL QSAKPALDLN 540
541 VSILASTFWP IDLSPHKIKC NFPKVLLAQI DQFTDFYLSK HTGRKLLWYP SMGSADVRVN 600
601 FKDRKYDLNV STIASVILLL FQDLKENQCL IFEEILEKTN IEVGDLKRNL QSLACAKYKI 660
661 LLKDPKGREV NAGDKFYFNE NFVSNLARIK ISTVAQTRVE DDSERKRTLE KVDESRKHQA 720
721 DACIVRVMKD RKVCEHNQLM AEVTRQLNPR FHPSPMMIKR RIEALIEREY LQRQADNGRI 780
781 YEYLA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
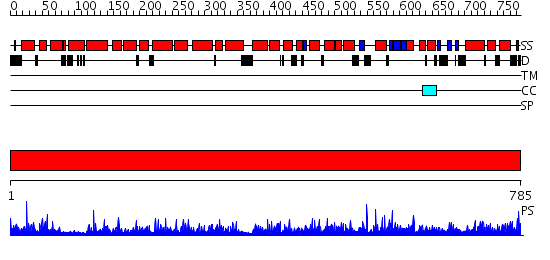
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..785] | 1000.0 | No description for 2hyeC was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)