













| General Information: |
|
| Name(s) found: |
cdc5 /
SPAC644.12
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 757 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA spliceosomal complex [IDA |
| Biological Process: |
RNA splicing, via transesterification reactions
[ISS G2/M transition of mitotic cell cycle [IMP nuclear mRNA splicing, via spliceosome [IMP |
| Molecular Function: |
DNA binding
[IEA]
protein binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVLKGGAWK NTEDEILKAA VSKYGKNQWA RISSLLVRKT PKQCKARWYE WIDPSIKKTE 60
61 WSREEDEKLL HLAKLLPTQW RTIAPIVGRT ATQCLERYQK LLDDLEAKEN EQLGLISGEG 120
121 AEAAAPVNDP NSRLRFGEAE PNLETLPALP DAIDMDEDEK EMLSEARARL ANTQGKKAKR 180
181 KDREKQLELT RRLSHLQKRR ELKAAGINIK LFRRKKNEMD YNASIPFEKK PAIGFYDTSE 240
241 EDRQNFREKR EADQKIIENG IRNNEMESEG RKFGHFEKPK PIDRVKKPNK DAQEEKMRRL 300
301 AEAEQMSKRR KLNLPSPTVS QDELDKVVKL GFAGDRARAM TDTTPDANYS TNLLGKYTQI 360
361 ERATPLRTPI SGELEGREDS VTIEVRNQLM RNREQSSLLG QESIPLQPGG TGYTGVTPSH 420
421 AANGSALAAP QATPFRTPRD TFSINAAAER AGRLASEREN KIRLKALREL LAKLPKPKND 480
481 YELMEPRFAD ETDVEATVGV LEEDATDRER RIQERIAEKE RLAKARRSQV IQRDLIRPSV 540
541 TQPEKWKRSL ENEDPTANVL LKEMIALISS DAINYPFGNS KVKGTANKVP DLSNEEIERC 600
601 RLLLKKEIGQ LESDDYIQFE KEFLETYSAL HNTSSLLPGL VIYEEDDEDV EAAEKFYTND 660
661 IQRDLAKKAL ECNKLENRVY DLVRSSYEQR NFLIKKISHA WKALQTERKN LTCYEFLYNQ 720
721 ERLALPNRLE AAEIELSKMQ QIEAYAQQDY ARVTGQN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
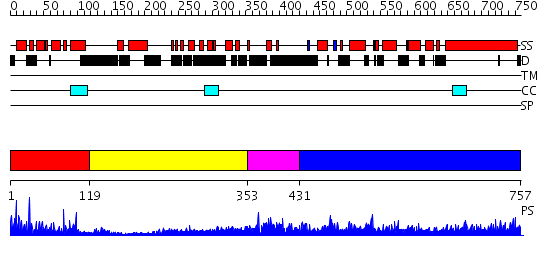
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | 33.69897 | c-Myb, DNA-binding domain | |
| 2 | View Details | [119..352] | 3.522879 | No description for 2i1jA was found. | |
| 3 | View Details | [353..430] | 1.120996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [431..757] | 5.69897 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)