













| General Information: |
|
| Name(s) found: |
FBpp0305074
[FlyBase]
FBpp0305073 [FlyBase] FBpp0297199 [FlyBase] ari-1-PC / FBpp0089023 [FlyBase] ari-1-PB / FBpp0088500 [FlyBase] ari-1-PA / FBpp0088499 [FlyBase] |
| Description(s) found:
Found 69 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 503 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[NAS]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
ubiquitin-protein ligase activity
[NAS]
zinc ion binding [IEA] protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSDNDNDFC DNVDSGNVSS GDDGDDDFGM EVDLPSSADR QMDQDDYQYK VLTTDEIVQH 60
61 QREIIDEANL LLKLPTPTTR ILLNHFKWDK EKLLEKYFDD NTDEFFKCAH VINPFNATEA 120
121 IKQKTSRSQC EECEICFSQL PPDSMAGLEC GHRFCMPCWH EYLSTKIVAE GLGQTISCAA 180
181 HGCDILVDDV TVANLVTDAR VRVKYQQLIT NSFVECNQLL RWCPSVDCTY AVKVPYAEPR 240
241 RVHCKCGHVF CFACGENWHD PVKCRWLKKW IKKCDDDSET SNWIAANTKE CPRCSVTIEK 300
301 DGGCNHMVCK NQNCKNEFCW VCLGSWEPHG SSWYNCNRYD EDEAKTARDA QEKLRSSLAR 360
361 YLHYYNRYMN HMQSMKFENK LYASVKQKME EMQQHNMSWI EVQFLKKAVD ILCQCRQTLM 420
421 YTYVFAYYLK KNNQSMIFED NQKDLESATE MLSEYLERDI TSENLADIKQ KVQDKYRYCE 480
481 KRCSVLLKHV HEGYDKEWWE YTE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
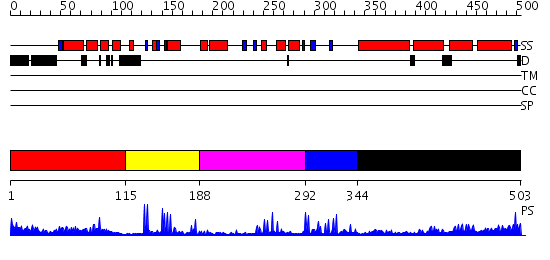
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..114] | 3.376994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [115..187] | 3.39794 | Solution Structure of the RING domain of the Tripartite motif protein 32 | |
| 3 | View Details | [188..291] | 3.4 | Solution Structure of the IBR domain of the RING finger protein 31 protein | |
| 4 | View Details | [292..343] | 8.221849 | Solution Structure of the C-terminal RING from a RING-IBR-RING (TRIAD) motif | |
| 5 | View Details | [344..503] | 7.302997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)