













| General Information: |
|
| Name(s) found: |
hep-PA /
FBpp0073578
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1178 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
dorsal closure
[IMP][NAS][TAS]
positive regulation of JUN kinase activity [IMP] dorsal closure, spreading of leading edge cells [IMP] determination of adult lifespan [IMP] JNK cascade [TAS] border follicle cell migration [IGI] protein amino acid phosphorylation [NAS][TAS] MAPKKK cascade [NAS] wound healing [IMP] response to heat [IMP] ovarian follicle cell development [IMP][TAS] basement membrane disassembly [IMP] triglyceride homeostasis [IMP] imaginal disc fusion, thorax closure [TAS] chorion-containing eggshell pattern formation [IMP] imaginal disc eversion [IMP] regulation of hemocyte differentiation [IMP] dorsal appendage formation [IMP] filopodium assembly [IMP] imaginal disc fusion [IMP] micropyle formation [IMP] regulation of transcription from RNA polymerase II promoter [NAS] axon extension [IMP] establishment of planar polarity [NAS] carbohydrate homeostasis [IMP] lamellipodium assembly [IMP] actin filament bundle assembly [IMP] |
| Molecular Function: |
ATP binding
[IEA]
receptor signaling protein serine/threonine kinase activity [NAS] protein binding [IPI] MAP kinase kinase activity [ISS][NAS][TAS] protein kinase activity [IDA] protein serine/threonine kinase activity [IEA] JUN kinase kinase activity [ISS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTIEFETIG SRLQSLEAKL QAQNESHDQI VLSGARGPVV SGSVPSARVP PLATSASAAT 60
61 SATHAPSLGA GSVSGSGISI AQRPAPPVPH ATPFGSASAS SSSSSASAFA SAAPATGTFG 120
121 GTYTPPTTRV SRATPTLPML SSGPGGGLNR TRPVILPLPT PPHPPVSETD MKLKIIMEQT 180
181 GKLNINGRQY PTDINDLKHL GDLGNGTSGN VVKMMHLSSN TIIAVKQMRR TGNAEENKRI 240
241 LMDLDVVLKS HDCKYIVKCL GCFVRDPDVW ICMELMSMCF DKLLKLSKKP VPEQILGKVT 300
301 VATVNALSYL KDKHGVIHRD VKPSNILIDE RGNIKLCDFG ISGRLVDSKA NTRSAGCAAY 360
361 MAPERIDPKK PKYDIRADVW SLGITLVELA TARSPYEGCN TDFEVLTKVL DSEPPCLPYG 420
421 EGYNFSQQFR DFVIKCLTKN HQDRPKYPEL LAQPFIRIYE SAKVDVPNWF QSIKDNRLRA 480
481 NGDPTLQRAT ATGSAIGSGA GSLAGSGSGS AGGAVKYGRA TTYAGQSPTN PQKTIKPTQI 540
541 PSYQQQQSQF FMQSATQLPQ TTTTTPTATT NCFGGSGNGN GRGNGSGGSG NGSGSSSSAS 600
601 PLSPPSAGIG DLNRLYRKSP FMQRKLSNGS HHPHYKYNDE SPKKESMFSS IGQSILRNLT 660
661 TSPFSQKKHN STATTIPLPH NNQTLITDAA TAAAAAATAT TPPNIAATVL TTTPTTTPTW 720
721 RLPTENSQAY DSCDSSSNAT TTTLNLGLSS PSPSLPRKQF PTESPTLQLT SQQQQQPQRL 780
781 QPGNQSPIVL QRFYHQQNQL REKEAAERYQ QQRQQPPVGV TSTNPFHSNY VPPPPSTHST 840
841 SSQSSTQSTC SQIAINPASI SPSSGSGTGN MAGLGIGSAP ASGLGAAGHF GAGGTGEQLQ 900
901 YQPLPIAAEA TGTSPTLQSR SPEQQSDYGG NGNMVASKLS KLYARRQLLG QSSSSGASNS 960
961 SLDGCSREQH DAGWFNTLAG AMKRQFATYV KTQLNSTATS PVASSMDRDQ EPVHPQPPAY1020
1021 RSVVNNGSGG KSYYYRTLSA ASSSSNTSQS TSPTTEPLPG GGTSSFLRRY ASSGPGGSIS1080
1081 TPPSPHILAG LDRRHRSPDP PPRYNRGQSP LLLRKNLLEL SGQPPGSPLL HRRYVSASPP1140
1141 LPPPRRGSES VPGSPQHFRT RIHYTPEPQR RIYRTIDQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
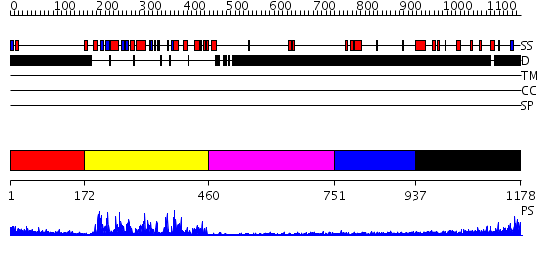
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 63.69897 | Abl tyrosine kinase, SH3 domain; Abl tyrosine kinase; Abelsone tyrosine kinase (abl) | |
| 2 | View Details | [172..459] | 61.221849 | No description for 2dylA was found. | |
| 3 | View Details | [460..750] | 1.01 | CRYSTAL STRUCTURE OF THE COLLAGEN-BINDING DOMAIN OF YERSINIA ADHESIN YadA | |
| 4 | View Details | [751..936] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [937..1178] | 2.314996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)