













| General Information: |
|
| Name(s) found: |
Gtp-bp-PA /
FBpp0073336
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 614 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum membrane
[IDA]
lipid particle [IDA] signal recognition particle receptor complex [ISS] |
| Biological Process: |
SRP-dependent cotranslational protein targeting to membrane
[IEA]
regulation of protein secretion [IEP] axonogenesis [IEP] |
| Molecular Function: |
GTP binding
[IEA][NAS]
protein binding [IEA] GTPase activity [IEA] mRNA binding [NAS] signal recognition particle binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLDFVVIFTK GGVVLWHSNA SGNSFASCIN SLIRGVILEE RNTEAKYYEE DHLAVQFKLD 60
61 NELDLVYAAI FQKVIKLNYL DGFLADMQAA FKEKYGDIRL GDDYDFDREY RRVLSAAEEA 120
121 SAKQVKAPKT MRSYNESQKS KKTVASMIQD DKKPVEKRVN IQEAPPPSKS QPSSPPTGSP 180
181 MDKIIMEKRR KLREKLTPTK KTSPSDSKSS KPEKAGKKPR VWDLGGNSKD AALLDRSRDS 240
241 PDDVQYQNIN SELVGTMQGV IRDLDVESED EADNEDASSE GEAEEQVQSK KGKRGGLLSY 300
301 FKGIVGAKTM SLADLQPALE KMRDHLISKN VASEIAAKLC DSVAASLDGK QMGTFDSIAS 360
361 QVKEALTESL VRILSPKRRI DIIRDALESK RNGRPYTIIF CGVNGVGKST NLAKICFWLI 420
421 ENDFNVLIAA CDTFRAGAVE QLRTHTRHLN ALHPAAKHDG RNMVQLYEKG YGKDAAGIAM 480
481 EAIKFAHDTR VDVVLVDTAG RMQDNEPLMR SLSKLIKVNN PDLVLFVGEA LVGNEAVDQL 540
541 VKFNQSLADY SSNENPHIID GIVLTKFDTI DDKVGAAISM TYITGQPIVF VGTGQTYADL 600
601 KAINVNAVVN SLMK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
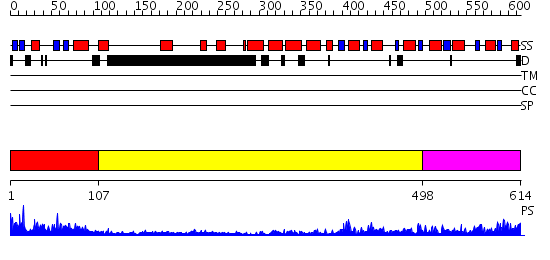
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | 39.522879 | The Structure of the Mammalian SRP Receptor | |
| 2 | View Details | [107..497] | 28.522879 | Crystal Structure Analysis of ClpB | |
| 3 | View Details | [498..614] | 62.69897 | No description for 2j289 was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)