













| General Information: |
|
| Name(s) found: |
ct-PA /
FBpp0071026
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2175 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][NAS]
|
| Biological Process: |
sensory organ boundary specification
[NAS]
spiracle morphogenesis, open tracheal system [IMP] regulation of gene-specific transcription [IMP] Malpighian tubule morphogenesis [NAS][TAS] imaginal disc-derived wing margin morphogenesis [IMP][NAS] peripheral nervous system development [TAS] regulation of transcription, DNA-dependent [IEA] female gonad development [TAS] antennal development [IEP][IMP] regulation of transcription [IEA] negative regulation of Notch signaling pathway [IMP] mitotic cell cycle [IMP] open tracheal system development [TAS] dendrite morphogenesis [IMP][TAS] sensory perception of sound [IMP] antennal joint development [IMP] ovarian follicle cell stalk formation [TAS] central nervous system development [TAS] oogenesis [IMP] sensory organ precursor cell fate determination [NAS] ovarian follicle cell development [IMP] cell fate commitment [NAS] |
| Molecular Function: |
DNA binding
[NAS][TAS]
transcription factor activity [IEA] specific RNA polymerase II transcription factor activity [NAS] sequence-specific DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQPTLPQAAG TADMDLTAVQ SINDWFFKKE QIYLLAQFWQ QRATLAEKEV NTLKEQLSTG 60
61 NPDSNLNSEN SDTAAAAATA AAVAAVVAGA TATNDIEDEQ QQQLQQTASG GILESDSDKL 120
121 LNSSIVAAAI TLQQQNGSNL LANTNTPSPS PPLLSAEQQQ QLQSSLQQSG GVGGACLNPK 180
181 LFFNHAQQMM MMEAAAAAAA AALQQQQQQQ SPLHSPANEV AIPTEQPAAT VATGAAAAAA 240
241 AAATPIATGN VKSGSTTSNA NHTNSNNSHQ DEEELDDEEE DEEEDEDEDD EEENASMQSN 300
301 ADDMELDAQQ ETRTEPSATT QQQHQQQDTE DLEENKDAGE ASLNVSNNHN TTDSNNSCSR 360
361 KNNNGGNESE QHVASSAEDD DCANNNTNTS NNNNTSNTAT SNTNNNNNNN SSSGNSEKRK 420
421 KKNNNNNNGQ PAVLLAAKDK EIKALLDELQ RLRAQEQTHL VQIQRLEEHL EVKRQHIIRL 480
481 EARLDKQQIN EALAEATALS AAASTNNNNN SQSSDNNKKL NTAAERPMDA SSNADLPEST 540
541 KAPVPAEDDE EDEDQAMLVD SEEAEDKPED SHHDDDEDED EDREAVNATT TDSNELKIKK 600
601 EQHSPLDLNV LSPNSAIAAA AAAAAAAACA NDPNKFQALL IERTKALAAE ALKNGASDAL 660
661 SEDAHHQQQQ HHQQQHQHQQ QHHQQQHLHQ QHHHHLQQQP NSGSNSNPAS NDHHHGHHLH 720
721 GHGLLHPSSA HHLHHQTTES NSNSSTPTAA GNNNGSNNSS SNTNANSTAQ LAASLASTLN 780
781 GTKSLMQEDS NGLAAVAMAA HAQHAAALGP GFLPGLPAFQ FAAAQVAAGG DGRGHYRFAD 840
841 SELQLPPGAS MAGRLGESLI PKGDPMEAKL QEMLRYNMDK YANQALDTLH ISRRVRELLS 900
901 VHNIGQRLFA KYILGLSQGT VSELLSKPKP WDKLTEKGRD SYRKMHAWAC DDNAVMLLKS 960
961 LIPKKDSGLP QYAGRGAGGA GGDDSMSEDR IAHILSEASS LMKQSSVAQH REQERRSHGG1020
1021 EDSHSNEDSK SPPQSCTSPF FKVENQLKQH QHLNPEQAAA QQREREREQR EREQQQRLRH1080
1081 DDQDKMARLY QELIARTPRE TAFPSFLFSP SLFGGAAGMP GAASNAFPAM ADENMRHVFE1140
1141 REIAKLQQHQ QQQQAAQAQA QFPNFSSLMA LQQQVLNGAQ DLSLAAAAAK DIKLNGQRSS1200
1201 LEHSAGSSSC SKDGERDDAY PSSLHGRKSE GGGTPAPPAP PSGPGTGAGA PPTAAPPTGG1260
1261 ASSNSAAPSP LSNSILPPAL SSQGEEFAAT ASPLQRMASI TNSLITQPPV TPHHSTPQRP1320
1321 TKAVLPPITQ QQFDMFNNLN TEDIVRRVKE ALSQYSISQR LFGESVLGLS QGSVSDLLAR1380
1381 PKPWHMLTQK GREPFIRMKM FLEDENAVHK LVASQYKIAP EKLMRTGSYS GSPQMPQGLA1440
1441 SKMQAASLPM QKMMSELKLQ EPAQAQHLMQ QMQAAAMSAA MQQQQVAQAQ QQAQQAQQAQ1500
1501 QHLQQQAQQH LQQQQHLAQQ QHPHQQHHQA AAAAAALHHQ SMLLTSPGLP PQHAISLPPS1560
1561 AGGAQPGGPG GNQGSSNPSN SEKKPMLMPV HGTNAMRSLH QHMSPTVYEM AALTQDLDTH1620
1621 DITTKIKEAL LANNIGQKIF GEAVLGLSQG SVSELLSKPK PWHMLSIKGR EPFIRMQLWL1680
1681 SDANNVERLQ LLKNERREAS KRRRSTGPNQ QDNSSDTSSN DTNDFYTSSP GPGSVGSGVG1740
1741 GAPPSKKQRV LFSEEQKEAL RLAFALDPYP NVGTIEFLAN ELGLATRTIT NWFHNHRMRL1800
1801 KQQVPHGPAG QDNPIPSRES TSATPFDPVQ FRILLQQRLL ELHKERMGMS GAPIPYPPYF1860
1861 AAAAILGRSL AGIPGAAAAA GAAAAAAAVG ASGGDELQAL NQAFKEQMSG LDLSMPTLKR1920
1921 ERSDDYQDDL ELEGGGHNLS DNESLEGQEP EDKTTDYEKV LHKSALAAAA AYMSNAVRSS1980
1981 RRKPAAPQWV NPAGAVTNPS AVVAAVAAAA AAAADNERII NGVCVMQASE YGRDDTDSNK2040
2041 PTDGGNDSDH EHAQLEIDQR FMEPEVHIKQ EEDDDEEQSG SVNLDNEDNA TSEQKLKVIN2100
2101 EEKLRMVRVR RLSSTGGGSS EEMPAPLAPP PPPPAASSSI VSGESTTSSS SSSNTSSSTP2160
2161 AVTTAAATAA AGWNY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
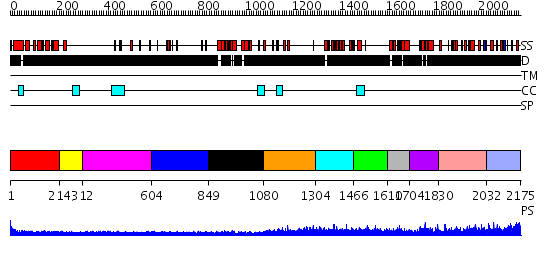
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..213] | 1.400995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [214..311] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [312..603] | 3.30103 | bI3 LAGLIDADG Maturase | |
| 4 | View Details | [604..848] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [849..1079] | 3.84 | Solution structure of the MAR-binding domain of SATB1 | |
| 6 | View Details | [1080..1303] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1304..1465] | 16.0 | Solution structure of the CUT domain of human homeobox protein Cux-2 (Cut-like 2) | |
| 8 | View Details | [1466..1609] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1610..1703] | 18.39794 | Solution structure of the third CUT domain of human Homeobox protein Cux-2 | |
| 10 | View Details | [1704..1829] | 16.39794 | Paired protein | |
| 11 | View Details | [1830..2031] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [2032..2175] | 1.046995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 9 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 10 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.62 |
Source: Reynolds et al. (2008)