













| General Information: |
|
| Name(s) found: |
dnc-PJ /
FBpp0070489
[FlyBase]
dnc-PI / FBpp0070487 [FlyBase] |
| Description(s) found:
Found 40 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1070 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
nonassociative learning
[TAS]
signal transduction [IEA] cyclic nucleotide metabolic process [NAS] synaptic transmission [IMP] olfactory learning [TAS] behavioral response to ethanol [NAS] learning [NAS] circadian rhythm [TAS] associative learning [TAS] memory [TAS] behavior [TAS] reproduction [NAS] conditioned taste aversion [IMP] learning or memory [NAS][TAS] mating behavior [TAS] short-term memory [IMP] oogenesis [TAS] locomotor rhythm [NAS] axon extension [IMP] courtship behavior [TAS] cAMP-mediated signaling [NAS] |
| Molecular Function: |
3',5'-cyclic-AMP phosphodiesterase activity
[ISS][TAS]
3',5'-cyclic-nucleotide phosphodiesterase activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQESNGGPA AGGGAAAAPP PPPQYIITTP SEVDPDEVRS MADLELGSPE KQVQVQSQKF 60
61 SSTSSTTKVA THSFSMSSSA GTTGQQSKQD SAQQIQQLQQ LQQLQQLQQQ QQQQQSQRII 120
121 SSSTRSQSLQ SSTIVGEATT ITSGAAQILS ASAAASLAQQ LKAQSSTSII TSSEQRTSTS 180
181 TSSSSSTRYI ASGSSNLAGG NSNSASSASS KTRFQSFLQQ PEGAHGFLTA HQKHVRQFVR 240
241 STSAHSEAAA GVAGARAEKC IRSASTQIDD ASVAGVVESA GNLTDSSATG GSMQLSMSKL 300
301 GLQQSSSILI SKSAETIEMK SSSAGMRTQL TLSGGFLAPP GNRKITILSP IHAPPGLHDM 360
361 LKRAQGRSPL SPRISFPGSD SDLFGFDVEN GQGARSPLEG GSPSAGLVLQ NLPQRRESFL 420
421 YRSDSDFEMS PKSMSRNSSI ASERFKEQEA SILVDRSHGE DLIVTPFAQI LASLRSVRNN 480
481 LLSLTNVPAS NKSRRPNQSS SASRSGNPPG APLSQGEEAY TRLATDTIEE LDWCLDQLET 540
541 IQTHRSVSDM ASLKFKRMLN KELSHFSESS RSGNQISEYI CSTFLDKQQE FDLPSLRVED 600
601 NPELVAANAA AGQQSAGQYA RSRSPRGPPM SQISGVKRPL SHTNSFTGER LPTFGVETPR 660
661 ENELGTLLGE LDTWGIQIFS IGEFSVNRPL TCVAYTIFQS RELLTSLMIP PKTFLNFMST 720
721 LEDHYVKDNP FHNSLHAADV TQSTNVLLNT PALEGVFTPL EVGGALFAAC IHDVDHPGLT 780
781 NQFLVNSSSE LALMYNDESV LENHHLAVAF KLLQNQGCDI FCNMQKKQRQ TLRKMVIDIV 840
841 LSTDMSKHMS LLADLKTMVE TKKVAGSGVL LLDNYTDRIQ VLENLVHCAD LSNPTKPLPL 900
901 YKRWVALLME EFFLQGDKER ESGMDISPMC DRHNATIEKS QVGFIDYIVH PLWETWADLV 960
961 HPDAQDILDT LEENRDYYQS MIPPSPPPSG VDENPQEDRI RFQVTLEESD QENLAELEEG1020
1021 DESGGESTTT GTTGTTAASA LSGAGGGGGG GGGMAPRTGG CQNQPQHGGM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
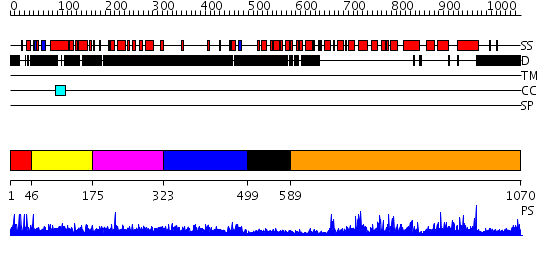
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..45] | 1.005992 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [46..174] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [175..322] | 1.027962 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [323..498] | 4.122973 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [499..588] | 1.220989 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [589..1070] | 143.0 | No description for 2fm0A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)