













| General Information: |
|
| Name(s) found: |
heph-PU /
FBpp0289497
[FlyBase]
heph-PT / FBpp0289496 [FlyBase] heph-PS / FBpp0289495 [FlyBase] heph-PC / FBpp0085239 [FlyBase] |
| Description(s) found:
Found 43 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 581 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
ribonucleoprotein complex [IDA] cytoplasm [IDA] |
| Biological Process: |
negative regulation of oskar mRNA translation
[IMP]
Notch signaling pathway [IMP] spermatid development [IMP] imaginal disc-derived wing margin morphogenesis [IMP] oocyte microtubule cytoskeleton polarization [IMP] imaginal disc-derived wing vein morphogenesis [IMP] nuclear mRNA splicing, via spliceosome [ISS] |
| Molecular Function: |
nucleotide binding
[IEA]
poly-pyrimidine tract binding [ISS][NAS] translation repressor activity, nucleic acid binding [IMP] mRNA binding [ISS] mRNA 3'-UTR binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRGSDELLSQ AAVMAPASDN NNQDLATKKA KLEPGTVLAG GIAKASKVIH LRNIPNESGE 60
61 ADVIALGIPF GRVTNVLVLK GKNQAFIEMA DEISATSMVS CYTVTPPQMR GRMVYVQFSN 120
121 HRELKTDQGH NNSTAHSDYS VQSPASGSPL PLSAAANATS NNANSSSDSN SAMGILQNTS 180
181 AVNAGGNTNA AGGPNTVLRV IVESLMYPVS LDILHQIFQR YGKVLKIVTF TKNNSFQALI 240
241 QYPDANSAQH AKSLLDGQNI YNGCCTLRID NSKLTALNVK YNNDKSRDFT NPALPPGEPG 300
301 VDIMPTAGGL MNTNDLLLIA ARQRPSLSGD KIVNGLGAPG VLPPFALGLG TPLTGGYNNA 360
361 LPNLAAFSLA NSGALQTTAP AMRGYSNVLL VSNLNEEMVT PDALFTLFGV YGDVQRVKIL 420
421 YNKKDSALIQ MAEPQQAYLA MSHLDKLRLW GKPIRVMASK HQAVQLPKEG QPDAGLTRDY 480
481 SQNPLHRFKK PGSKNYQNIY PPSATLHLSN IPSSCSEDDI KEAFTSNSFE VKAFKFFPKD 540
541 RKMALLQLLS VEEAVLALIK MHNHQLSESN HLRVSFSKSN I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
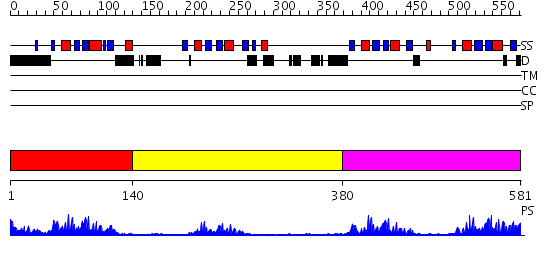
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..139] | 16.0 | Solution structure of Polypyrimidine Tract Binding protein RBD1 complexed with CUCUCU RNA | |
| 2 | View Details | [140..379] | 11.522879 | Solution structure of Polypyrimidine Tract Binding protein RBD2 complexed with CUCUCU RNA | |
| 3 | View Details | [380..581] | 39.522879 | Solution structure of Polypyrimidine Tract Binding protein RBD34 complexed with CUCUCU RNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)