













| General Information: |
|
| Name(s) found: |
heph-PR /
FBpp0289494
[FlyBase]
heph-PO / FBpp0289491 [FlyBase] heph-PH / FBpp0085240 [FlyBase] |
| Description(s) found:
Found 36 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 789 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
ribonucleoprotein complex [IDA] cytoplasm [IDA] |
| Biological Process: |
negative regulation of oskar mRNA translation
[IMP]
Notch signaling pathway [IMP] spermatid development [IMP] imaginal disc-derived wing margin morphogenesis [IMP] oocyte microtubule cytoskeleton polarization [IMP] imaginal disc-derived wing vein morphogenesis [IMP] nuclear mRNA splicing, via spliceosome [ISS] |
| Molecular Function: |
nucleotide binding
[IEA]
poly-pyrimidine tract binding [ISS][NAS] translation repressor activity, nucleic acid binding [IMP] mRNA binding [ISS] mRNA 3'-UTR binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMSCPIPMPM PISSLHKSEF EPVNIGVKQM QLLHAPASTH PHPHPHQHPQ HSAASAAAYQ 60
61 AMQVHQQQHQ QQQQAAVFHH QVAAIAAQQQ QQQQQAQVQH QQQMQQQFHY QAQAQAQAQA 120
121 MLQTYMQLQV NSAVGVSTAP ATPTKPVAAQ IPGAISGPIA GPVPVSVPTQ FFTAPSTPLT 180
181 IIPAAAVASS AMFATAATPT SAATTPTNNR GSDELLSQAA VMAPASDNNN QDLATKKAKL 240
241 EPGTVLAGGI AKASKVIHLR NIPNESGEAD VIALGIPFGR VTNVLVLKGK NQAFIEMADE 300
301 ISATSMVSCY TVTPPQMRGR MVYVQFSNHR ELKTDQGHNN STAHSDYSVQ SPASGSPLPL 360
361 SAAANATSNN ANSSSDSNSA MGILQNTSAV NAGGNTNAAG GPNTVLRVIV ESLMYPVSLD 420
421 ILHQIFQRYG KVLKIVTFTK NNSFQALIQY PDANSAQHAK SLLDGQNIYN GCCTLRIDNS 480
481 KLTALNVKYN NDKSRDFTNP ALPPGEPGVD IMPTAGGLMN TNDLLLIAAR QRPSLSGDKI 540
541 VNGLGAPGVL PPFALGLGTP LTGGYNNALP NLAAFSLANS GALQTTAPAM RGYSNVLLVS 600
601 NLNEEMVTPD ALFTLFGVYG DVQRVKILYN KKDSALIQMA EPQQAYLAMS HLDKLRLWGK 660
661 PIRVMASKHQ AVQLPKEGQP DAGLTRDYSQ NPLHRFKKPG SKNYQNIYPP SATLHLSNIP 720
721 SSCSEDDIKE AFTSNSFEVK AFKFFPKDRK MALLQLLSVE EAVLALIKMH NHQLSESNHL 780
781 RVSFSKSNI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
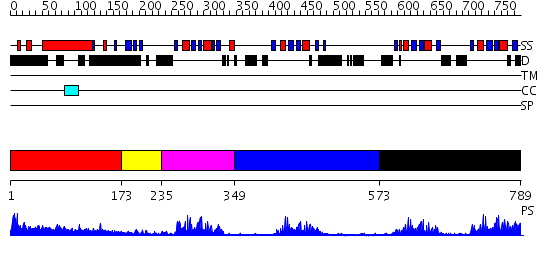
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..172] | 3.154902 | Sec24 | |
| 2 | View Details | [173..234] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [235..348] | 15.30103 | Solution structure of Polypyrimidine Tract Binding protein RBD1 complexed with CUCUCU RNA | |
| 4 | View Details | [349..572] | 17.30103 | Solution structure of Polypyrimidine Tract Binding protein RBD2 complexed with CUCUCU RNA | |
| 5 | View Details | [573..789] | 44.0 | Solution structure of Polypyrimidine Tract Binding protein RBD34 complexed with CUCUCU RNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)