













| General Information: |
|
| Name(s) found: |
FBpp0307959
[FlyBase]
Med-PA / FBpp0085176 [FlyBase] |
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 771 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IPI][TAS]
transcription factor complex [IEA] cytoplasm [IDA] |
| Biological Process: |
cell proliferation
[IMP]
imaginal disc-derived leg morphogenesis [IMP] negative regulation of salivary gland boundary specification [TAS] retinal ganglion cell axon guidance [IMP] transforming growth factor beta receptor signaling pathway [IGI][IMP][ISS][TAS] imaginal disc-derived wing morphogenesis [IMP] germ-line stem cell division [IMP][TAS] regulation of transcription, DNA-dependent [IEA][NAS] dorsal/ventral axis specification [IMP] neuron development [IMP] positive regulation of synaptic growth at neuromuscular junction [IMP] dorsal/ventral pattern formation [IMP] heart development [TAS] germ-line stem cell maintenance [IMP] BMP signaling pathway [TAS] ovarian follicle cell development [IMP] somatic stem cell maintenance [IMP] |
| Molecular Function: |
DNA binding
[TAS]
transcription factor activity [IEA] protein binding [IPI] RNA polymerase II transcription factor activity [NAS] transforming growth factor beta receptor, common-partner cytoplasmic mediator activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGGGSGACPP AHMYGAVAPQ DIIVRDMVQM PPPPSNAPTS ADACLSIVHS LMCHRQGGES 60
61 EGFAKRAIES LVKKLKEKRD ELDSLITAIT TNGAHPSKCV TIQRTLDGRL QVAGRKGFPH 120
121 VIYARIWRWP DLHKNELKHV KYCAFAFDLK CDSVCVNPYH YERVVSPGID LSGLSLQSGP 180
181 SRLVKDEYSA GPLVGSMDID GNDIGTIQHH PTQMVGPGGY GYPQGPSEYV GDANPMSAMF 240
241 PTGRTIPKIE PQDGVAGSRG SWMVPPPPRL GQPPQQQQQQ PQQQTPQPTQ QQQAQSQAAA 300
301 HSLPVPHGMP GMPGPMNPGP VMAPPPPPQQ AQNPQGNGVH HTQANSPTDP ASALAMQQQQ 360
361 QQQQQQQQQQ QQQQQQSGGV PNGSVNAGGG AAAGGQYYGQ PPPVSQMQGA GGGGTSVAPS 420
421 VHAQQNGYVS QPGSAGSAPV GGGGVFGTAQ PTPQQPQQPP TGVQANTGSA GAQAGAGGGA 480
481 AGTWTGPNTL TYTQSMQPPD PRSLPGGFWN SSLSGDLGSP QQTPPQQQQQ QQQPRLLSRQ 540
541 PPPEYWCSIA YFELDTQVGE TFKVPSAKPN VIIDGYVDPS GGNRFCLGAL SNVHRTEQSE 600
601 RARLHIGKGV QLDLRGEGDV WLRCLSDNSV FVQSYYLDRE AGRTPGDAVH KIYPAACIKV 660
661 FDLRQCHQQM HSLATNAQAA AAAQAAAVAG VANQQMGGGG RSMTAAAGIG VDDLRRLCIL 720
721 RLSFVKGWGP DYPRQSIKET PCWIEVHLHR ALQLLDEVLH AMPIDGPRAA A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
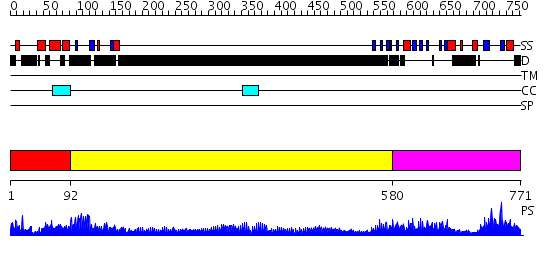
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] | 38.522879 | Crystal structure of Smad3-MH1 bound to DNA at 2.4 A resolution | |
| 2 | View Details | [92..579] | 41.0 | No description for 1y0fB was found. | |
| 3 | View Details | [580..771] | 95.221849 | Crystal Structure of the phosphorylated Smad2/Smad4 heterotrimeric complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)