













| General Information: |
|
| Name(s) found: |
Sirt7-PA /
FBpp0084733
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 771 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein amino acid deacetylation
[IEA]
chromatin silencing [IEA] regulation of transcription [IEA] |
| Molecular Function: |
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides
[IEA]
zinc ion binding [IEA] NAD binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKDLGEEKD QDQEQNTEME PKQEMDVAQS YITRAKMNPA KKDNEKRRRK DAMRRVSMIL 60
61 RKCDSMRTTE DRQFLEKHPD MVKTTKKRKE RVEIYKERVV EREDAPHVIE AKVEQLANII 120
121 SQAKHLVCYT GAGISTAALI PDYRGSQGIW TLLQKGQDIG EHDLSSANPT YTHMALYELH 180
181 RRRLLHHVVS QNCDGLHLRS GLPRNSLSEI HGNMYVEVCK NCRPNSVYWR QFDTTEMTAR 240
241 YCHKTHRLCH RCSEPLYDTI VHFGERGNVK WPLNWAGATA NAQRADVILC LGSSLKVLKK 300
301 YTWLWQMDRP ARQRAKICVV NLQWTPKDAI ASIKINGKCD QVMAQLMHLL HIPVPVYTKE 360
361 KDPIFAHASL LMPEELHTLT QPLLKNADEE EAFTTTTEET QDSTISSESC SFNYSDLPIG 420
421 KGPRIRTPIK NGRRVKTNLE LRQKFKTLNG QDEEIKVEHV KTNGEVKTEK DLITLESSIK 480
481 IETEVKLEKL ECSDTNFQQE LKLLELLPKL EPLSLKEETE ETPSNGFPEL PKLVAIQKTH 540
541 AECLSAVPTE SRLKPLQLPP LVPIGAPLST PFVEPKLVLP PASQSSSIQI KSEGDGDSST 600
601 ENDNEEEEES ELAQMDLLRQ NNDEELLRQL PTWYDAKYAY SGLHSILIPP PADLNIWNSQ 660
661 VVPNFAMNRS AASCFFCFDR YAELECQFYR RWNLSQRKHK KRARSGRFVV CECCPTSDDD 720
721 DDYDENISLA HIAAAETAKR RQQLSTSFPR KLARTQAGWY GKGYKKGRKR R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
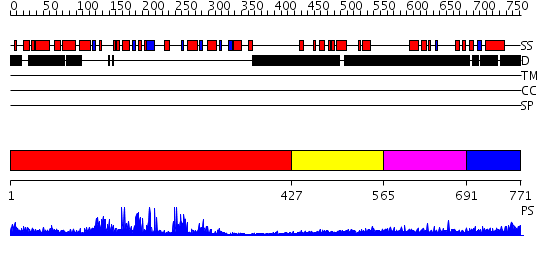
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..426] | 58.0 | Sirt2 histone deacetylase | |
| 2 | View Details | [427..564] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [565..690] | 1.202996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [691..771] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)