













| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFKCIPIFKG CNRQVEFVDK RHCSLPQVPE EILRYSRTLE ELFLDANHIR DLPKNFFRLH 60
61 RLRKLGLSDN EIGRLPPDIQ NFENLVELDV SRNDIPDIPD DIKHLQSLQV ADFSSNPIPK 120
121 LPSGFSQLKN LTVLGLNDMS LTTLPADFGS LTQLESLELR ENLLKHLPET ISQLTKLKRL 180
181 DLGDNEIEDL PPYLGYLPGL HELWLDHNQL QRLPPELGLL TKLTYLDVSE NRLEELPNEI 240
241 SGLVSLTDLD LAQNLLEALP DGIAKLSRLT ILKLDQNRLQ RLNDTLGNCE NMQELILTEN 300
301 FLSELPASIG QMTKLNNLNV DRNALEYLPL EIGQCANLGV LSLRDNKLKK LPPELGNCTV 360
361 LHVLDVSGNQ LLYLPYSLVN LQLKAVWLSE NQSQPLLTFQ PDTDAETGEQ VLSCYLLPQQ 420
421 EYQPITPARD LESDSEPFEE REPSRTVVKF SEEATQEKET PFVRQNTPHP KDLKAKAQKL 480
481 KVERSRNEEH ANLVTLPEEN GTKLAETPTE TRTIANNHQQ QPHPVQQPIV GVNSKQPVVV 540
541 GVVTPTTTTV APTGVQGGSE GASSTANNVK AATAAVVAEL AATVGGSDEV QDDDEQEDEF 600
601 ESDRRVGFQV EGEDDDFYKR PPKLHRRDTP HHLKNKRVQH LTDKQASEIL ANALASQERN 660
661 DTTPQHSLSG KVTSPIEEEE QLEVEQEQQQ QQQQHPFDSS LSPISAGKTA EASTDPDNLD 720
721 GVTELRLEQY EIHIERTAAG LGLSIAGGKG STPFKGDDDG IFISRVTEAG PADLAGLKVG 780
781 DKVIKVNGIV VVDADHYQAV QVLKACGAVL VLVVQREVTR LIGHPVFSED GSVSQISVET 840
841 RPLVADAPPA ASISHERYIP APIEIVPQQQ HLQQQQQQPI QQVAPTHSYS GNVFATPTAA 900
901 QTVQPAVSAA PNGLLLNGRE APLSYIQLHT TLIRDQIGQG LGFSIAGGKG SPPFKDDCDG 960
961 IFISRITEGG LAYRDGKIMV GDRVMAINGN DMTEAHHDAA VACLTEPQRF VRLVLQREYR1020
1021 GPLEPPTSPR SPAVLNSLSP SGYLANRPAN FSRSVVEVEQ PYKYNTLATT TPTPKPTVPA1080
1081 SISNNNNTLP SSKTNGFATA AAATIDSSTG QPVPAPRRTN SVPMGDGDIG AGSTTSGDSG1140
1141 EAQPSSLRPL TSDDFQAMIP AHFLSGGSQH QVHVARPNEV GVSAVTVNVN KPQPDLPMFP1200
1201 AAPTELGRVT ETITKSTFTE TVMTRITDNQ LAEPLISEEV VLPKNQGSLG FSIIGGTDHS1260
1261 CVPFGTREPG IFISHIVPGG IASKCGKLRM GDRILKVNEA DVSKATHQDA VLELLKPGDE1320
1321 IKLTIQHDPL PPGFQEVLLS KAEGERLGMH IKGGLNGQRG NPADPSDEGV FVSKINSVGA1380
1381 ARRDGRLKVG MRLLEVNGHS LLGASHQDAV NVLRNAGNEI QLVVCKGYDK SNLIHSIGQA1440
1441 GGMSTGFNSS ASCSGGSRQG SRASETGSEL SQSQSVSSLD HEEDERLRQD FDVFASQKPD1500
1501 AQQPTGPSVL AAAAAMVHGA SSPTPPAATS NITPLPTAAA VASADLTAPD TPATQTVALI1560
1561 HAEQQAHQQQ QQTQLAPLGQ EKSTQEKVLE IVRAADAFTT VPPKSPSEHH EQDKIQKTTT1620
1621 VVISKHTLDT NPTTPTTPAA PLSIAGAESA NSAGAPSPAV PASTPGSAPV LPAVAVQTQT1680
1681 QTTSTEKDEE EESQLQSTPA SRDGAEEQQE EVRAKPTPTK VPKSVSDKKR FFESAMEDQH1740
1741 KPTQKTDKVF SFLSKDEVEK LRQEEERKIA TLRRDKNSRL LDAANDNIDK DAAQQRTKSN1800
1801 SNSSSGDDND DSDQEEGIAR GDSVDNAALG HFDDAEDMRN PLDEIEAVFR S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
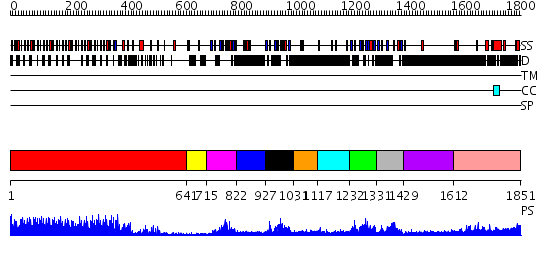
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..640] | 70.045757 | The molecular structure of toll-like receptor 3 ligand binding domain | |
| 2 | View Details | [641..714] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [715..821] | 15.154902 | Na+/H+ exchanger regulatory factor, NHERF | |
| 4 | View Details | [822..926] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [927..1030] | 23.0 | Solution structure of the first PDZ domain of scribble homolog protein (hScrib) | |
| 6 | View Details | [1031..1116] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1117..1231] | 25.0 | Structural Insights into the Inter-domain Chaperoning of Tandem PDZ Domains in Glutamate Receptor Interacting Proteins | |
| 8 | View Details | [1232..1330] | 4.83 | Solution structure of the alternatively spliced PDZ2 domain (PDZ2b) of PTP-Bas (hPTP1E) | |
| 9 | View Details | [1331..1428] | 21.39794 | Solution structure of the fourth PDZ domain of human scribble (KIAA0147 protein) | |
| 10 | View Details | [1429..1611] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1612..1851] | 1.136996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)