













| General Information: |
|
| Name(s) found: |
Eip93F-PA /
FBpp0083567
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1165 amino acids |
Gene Ontology: |
|
| Cellular Component: |
polytene chromosome
[NAS][TAS]
nucleus [NAS][TAS] |
| Biological Process: |
induction of apoptosis by hormones
[IMP]
larval midgut histolysis [TAS] phagocytosis, engulfment [TAS] positive regulation of transcription, DNA-dependent [TAS] autophagy [IMP] autophagic cell death [IEP][TAS] salivary gland cell autophagic cell death [IEP][NAS] ecdysone-mediated induction of salivary gland cell autophagic cell death [TAS] activation of caspase activity [IMP] regulation of transcription [NAS] |
| Molecular Function: |
DNA binding
[IEA][NAS]
transcription factor activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHISSYEISL ERVAEECMGR RQWKHYQDKL TCSHLNIEEQ QPIAIAGSED EPSQYNHSSK 60
61 EISQSNPNHC KTENHRLEQQ HNGSQLLEEE DSENNQTSHD SSRTPTPGAT STPSPPPEPI 120
121 DWRPSAKCNF CVNGRLLTVN AQGKLVAESA ATATSSSTSN SHIHQHDSDS NSSASLPHHI 180
181 SSSSSSNNNS SGNRARHIAA ASARATPAAA TPANSLELYK LLTQRAAKMT SMDSMAAQLA 240
241 QFSLLADFNL INSLASQQQQ QQQQQIASAV TPTTSEVSAA AISPALKDTP SPSVDAPLDL 300
301 SSKPSPNSSI SGDVKSVRAC ATPTPSGRRA YSEEDLSRAL QDVVANKLDA RKSASQHHEQ 360
361 RSILDNRLFK MKHHDQEQDH DGDELEDSND DAEAEVDSNA STPVYPAEFA RAQLRKLSHL 420
421 SEHNGSDLGE DVDRGSPKMG RHPACGNASA NQGAPPSIPL DANVLLHTLM LAAGIGAMPK 480
481 LDETQTVGDF IKGLLVANSG GIMNEGLLNL LSASQENSNG NASLLLQQQQ HQQHHQQHHQ 540
541 QQQQQQHVAA YRHRLPKSET PETNSSLDPN DASEDPILKI PSFKVSGPAS SSSLSPGGLV 600
601 GGHHHPLNNN NSLSISNNSN HSSNSHRNGS NRSPHSASPM LAAAVAQGGY SAGNSLLTSS 660
661 SSSIQKMMAS NIQRQINEQS GQESLRNGNV SDCSSNNGGS SSLGYKKPSI SVAKIIGGTD 720
721 TSRFGASPNL LSQQHHSAHH LTHQQQQQQL SAQEALGKGT RPKRGKYRNY DRDSLVEAVK 780
781 AVQRGEMSVH RAGSYYGVPH STLEYKVKER HLMRPRKREP KPQPDLVGLT GPANKLQLDK 840
841 LKAGPHGGSK LSNALKNQNN QAAAAAAAAA AAAAAATPNG LKLPLFEAGP QALSFQPNMF 900
901 WPQTNATNAY GLDFNRITEA MRNPQASNHH GLMKSAQDMV ENVYDGIIRK TLQASEGNGS 960
961 AAGNGSNGSN GNGHGHGHGH GHALLDQLLV KKTPLPFTNH RNNDYAATCS SASGESVKRS1020
1021 GSPMGNYADI KRERLSADSG GSSDEEHSAS HINNNNSDLA HNKNKSGGGG GGGGNGQTNG1080
1081 NGRSSRMTSR DDSETDASSL KSGESGGQQN HKMMDLNGGS SSSSHIKCES EAATGHHSPG1140
1141 HHTTSILHEK LAQIKAEQVD QADQL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
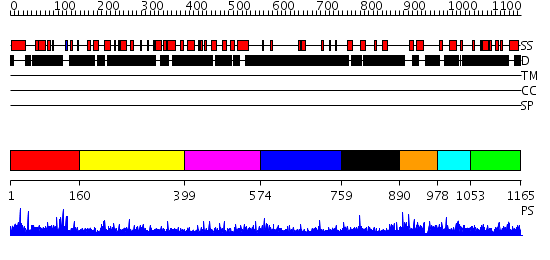
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..159] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [160..398] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [399..573] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [574..758] | 6.236997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [759..889] | 14.30103 | Solution structures of the HTH domain of human LCoR protein | |
| 6 | View Details | [890..977] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [978..1052] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1053..1165] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)