













| General Information: |
|
| Name(s) found: |
SNF4Agamma-PS /
FBpp0290804
[FlyBase]
SNF4Agamma-PR / FBpp0290803 [FlyBase] SNF4Agamma-PF / FBpp0083423 [FlyBase] |
| Description(s) found:
Found 38 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1400 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
positive regulation of cell cycle
[IGI]
protein amino acid phosphorylation [ISS] cholesterol homeostasis [IMP] |
| Molecular Function: |
AMP-activated protein kinase activity
[ISS][NAS]
protein serine/threonine kinase activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATNCDFIEQ PVKQQPLANA PVMGVQEDKA QEPKELPDEP AKQPDGQPYQ HFAGEDSFEC 60
61 AFESTMDDLE QGVLDGYESD SEASANDQIT DSASSGSLGQ LAQQQQREER LRGLPGPLHG 120
121 QHIDSEMLKC MPRSNSFDFH SSSGSSSDDE DDPDDDDDDQ VHHSLGLSLS GKFVKVDLYS 180
181 GHQRPGYLLP PYIERRRLSQ CKEEDDEDEG DRTQLGGGSS STGESTKSEK ASKAPPRPPT 240
241 PQDEASKEKF KDLERLRQQF MHKIDRINTR NIEESEAPPP PPPPPPPIPA TILPKALKAL 300
301 LHTQEPVAPP SRPSPVGPPS PAPTSSVIVA KETPVPVTPE PVSPLPLPTE PPLIIKGKFK 360
361 VTLAQETPEL RVEAQKLSKL NASSNAHTIS FPSSFGGYSS VQGLFSQRYG SNRFPTAAPH 420
421 MSKQFFEPSL VEIRTPAQSN VALNTTGLAN SNHQSLTCDT NSLANKPRTP SERELDEIWI 480
481 RRQDGGLSGT AAPGAPGAPL PLLQQQQQQQ RPVSLGGAST MPRLLRSESS ASTTSPSRQL 540
541 TNSGSVSVSV SDDEQDGSLG GAGRMPGSRP SSAPAEPNAK AAKKAYKKME KEQRRQEKEQ 600
601 SRREKEARKL ERETARRNER EAAKLEKINR HSEKISRSTE RMAMAGVGSR SGSLERRRSG 660
661 EDSPVLNQST VHGIASPNRR PTIFDVFRPR AKSDAKRQKE KHLLDPSSAD SSATSSTYSV 720
721 SGGTGPATSS AAGGAGGAAA AAGQGAAGGA GGLMNSMKVA MQNFSHRQHP AVTITSADGT 780
781 QSTAKSKYKD GSAHPHQGSD AQYYHTVTAV RPNSSQRSPM TKVMDLFRHR SSSVVSEADK 840
841 RKARAAAHQQ QLAVQSAHMR RASADLEKRR ASVGAAGRGL RGDGTLDPHH AAILFRDSRG 900
901 LPVADPFLEK VNLSDLEEDD SQIFVKFFRF HKCYDLIPTS AKLVVFDTQL LVKKAFYALV 960
961 YNGVRAAPLW DSEKQQFVGM LTITDFIKIL QMYYKSPNAS MEQLEEHKLD TWRSVLHNQV1020
1021 MPLVSIGPDA SLYDAIKILI HSRIHRLPVI DPATGNVLYI LTHKRILRFL FLYINELPKP1080
1081 AYMQKSLREL KIGTYNNIET ADETTSIITA LKKFVERRVS ALPLVDSDGR LVDIYAKFDV1140
1141 INLAAEKTYN DLDVSLRKAN EHRNEWFEGV QKCNLDESLY TIMERIVRAE VHRLVVVDEN1200
1201 RKVIGIISLS DILLYLVLRP SGEGVGGSES SLRASDPVLL RKVAEVEIPA TAAAATTTTP1260
1261 PRSPSAGSGN RSLIEDIPEE ETAPARSDDA DSDNNKSASE DKANNNQHDQ TTTAATANGD1320
1321 SNNSPVEVSF ADEAQEEEAA DQVERSNCDD DDQPALAEIE RKNASMDDDE DDGMSSAVSA1380
1381 ASALGQSLTP AAQEMALVSE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
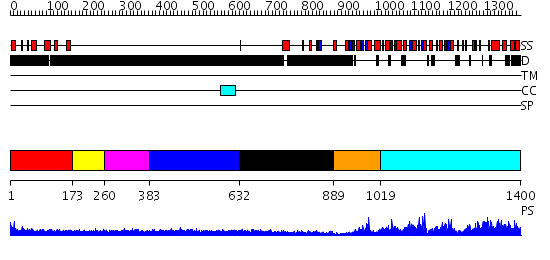
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..172] | 1.17399 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [173..259] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [260..382] | 2.196995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [383..631] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [632..888] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [889..1018] | 84.522879 | No description for 2v8qE was found. | |
| 7 | View Details | [1019..1400] | 125.0 | Inosine monophosphate dehydrogenase (IMPDH); Type II inosine monophosphate dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)