













| General Information: |
|
| Name(s) found: |
pnr-PA /
FBpp0082675
[FlyBase]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 540 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
|
| Biological Process: |
dorsal closure
[IMP][TAS]
heart contraction [IMP] cardiac cell differentiation [IMP] blastoderm segmentation [IMP] transforming growth factor beta receptor signaling pathway [TAS] pigment metabolic process [IMP] mesoderm development [NAS] pattern specification process [NAS] cardioblast cell fate determination [IMP] regulation of transcription, DNA-dependent [IEA] ectoderm development [TAS] embryonic heart tube development [IMP] regulation of transcription [ISS] pericardial cell differentiation [IMP] heart development [TAS] bristle morphogenesis [TAS] negative regulation of transcription, DNA-dependent [TAS] positive regulation of transcription, DNA-dependent [NAS] lymph gland development [IMP] cell differentiation [NAS] cardioblast differentiation [IMP] |
| Molecular Function: |
transcription factor activity
[IEA][ISS][TAS]
zinc ion binding [IEA] sequence-specific DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGILLSDGDS TSDQQSTRDY PHFSGDYQNV TLSAASASTS ASASATHVAA VKMYHSSAVA 60
61 AYTDLAAAGS AASAGVGVGV SGYHQQAVNA PVYVPSNRQY NHVAAHFGSA AAQNAWTTEG 120
121 FGSAHAQFYS PNAAVMMGSW RSAYDPSGFQ RSSPYESAMD FQFGEGRECV NCGAISTPLW 180
181 RRDGTGHYLC NACGLYHKMN GMNRPLIKPS KRLVSATATR RMGLCCTNCG TRTTTLWRRN 240
241 NDGEPVCNAC GLYYKLHGVN RPLAMRKDGI QTRKRKPKKT GSGSAVGAGT GSGTGSTLEA 300
301 IKECKEEHDL KPSLSLERHS LSKLHTDMKS GTSSSSTLMG HHSAQQQQQQ QQQQQQQQQQ 360
361 QQQQSAHQQC FPLYGQTTTQ QQHQQHGHSM TSSSGQAHLS ARHLHGAAGT QLYTPGSSSG 420
421 GGSASAYTSH SAETPALSNG TPSPHYQHHH HLGGTHGHHV TAAAAHHHFH AAAAVAAYGV 480
481 KTEASATNYD YVNNCYFGGT FGALGGAATT TAMAGGAASE LAGYHHQHNV IQAAKLMATS 540
541 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
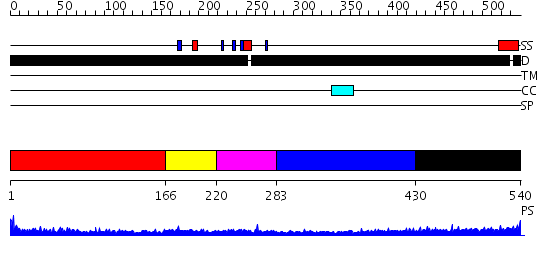
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..165] | 1.155991 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [166..219] | 2.68 | Erythroid transcription factor GATA-1 | |
| 3 | View Details | [220..282] | 10.39794 | Erythroid transcription factor GATA-1 | |
| 4 | View Details | [283..429] | 1.170991 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [430..540] | 7.162984 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)