













| General Information: |
|
| Name(s) found: |
gi|24647318
[NCBI NR]
gi|23171428 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1929 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
Toll signaling pathway
[IMP]
innate immune response [IMP] antimicrobial humoral response [IMP] positive regulation of antibacterial peptide biosynthetic process [IMP] |
| Molecular Function: |
ATP binding
[IEA]
DNA binding [IEA] helicase activity [NAS] ATP-dependent DNA helicase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLIVFNIPCR LDRLFILLET GSTAATRQAA AKQIGEVQKL YPHELHALLN RLVGYLHSTS 60
61 WETRIAAAQT VEAILKQVPP WQPEFQVLPK KERCATNTAT AAAAAEEDSC QSSGSNSTTA 120
121 SERLLSFDEF DLQQILHKGA RLIGSEGNEF DVQEEQPASG GGEEDRSLAS RLSRQRAVLN 180
181 DKLGLTTAAK LGVNLTDMIT DEDVALNRSG STYNVNAEKL PVEHLLNIKT AANGSGQAPL 240
241 SCREMNRAKR KARQNTSGCS VTAVTPTCSR RNSNSNHSTG SNHSSNGTTG AAIEEPEKKK 300
301 LKSTDRQEIF YSLNAAVPDA TGTWVDAVSW PLENFCSRLF IDLFHAKWEV RHGAATALRE 360
361 LINQHAQGAG KAVLQTREQM DEAHSRWLED AALRLLCVLC LDRFGDFVSD QVVAPVRETC 420
421 AQVLGTLVKE MEASKVQRIV NILLQLIQHK NEWDVRHGGL LGLKYVFVVR EELLPIYLPQ 480
481 SISNILIGLL DAVDDVGAVA AATLIPVSSW LPKLLNPAQV SSIVKMLWDL LLDQDELTSA 540
541 CNSFMGLLAA ILCLPNAARW VEMEPMAILV PRLWPFLSHS TSSVRRSTLK TLMTLTSADR 600
601 VKSEPKDENQ VKEEADKEEM KLNFGVSDWK WELLQQALRH IYQRILVEPQ ADIQALARQV 660
661 WSNLIQHADL GALLHAACPF VSSWICLSMQ PPRLAFDAGV LIRAGGDVST PGSTSRKRTP 720
721 RIGDELGGNI LAQSNASLKL YLGGSEATPL DVRDANFMRA RISSSRALGA LSHYLVQPAP 780
781 GVVYTPQTES PTDCYTKVLL GHLNAHSAVQ RIVCGLIIAF WALEDEPVRP GPPNLQEKLH 840
841 QCVSEYVYYD EVAISLTRLL QEAQDFIATL KQNKIPINDF NNAKILTLDQ IEAVATTLSE 900
901 NLRSYPLKPK VLDMLEERRR SLQASYQQTT SEQSAYHVSA QAALAGAICA LHCLPDKLNP 960
961 VVKPLMESIK REQCLQLQQL SAEFLVHLMD QVCDRNPSPN SKILNNLCTL LRSDPEFTPK1020
1021 LVMPLETLKQ TPVSSEVINN CVYYGILTLA LQQTVTTTNT RSGAGGATTP TATTAPRGPG1080
1081 RPPTGEILSA TNATAHIELK AQQAKEAEAK QCRIQRLGAA CAIEKLCRIF GEQIIEKVAV1140
1141 FQHLMFGKVE QFVKEAYDWR LGSLLPDLGV CNELVSSMQL IETAAPHLHV ALHPQMFALL1200
1201 PSLGFIVCHP LKAVRHMAAR CIAVLAEIDA CQTMQFVVHD LLPLLGKIEQ LIERQGAIEA1260
1261 IERVVSRLQI KVVPYIVLLV VPLLGAMSDP DESVRLLSTH CFANLVQLMP LDGKTEQLKS1320
1321 DPLQARKTRD REFLDYLFNP KSIPNYKVPV PISVELRCYQ QAGINWLWFL NKYNLHGILC1380
1381 DDMGLGKTLQ TICILAGDHM HRQTANLANL PSLVICPPTL TGHWVYEVEK FLDQGSVLRP1440
1441 LHYYGFPVGR EKLRSDIGTK CNLVVASYDT VRKDIDFFSG IHFNYCVLDE GHIIKNGKTK1500
1501 SSKAIKRLKA NHRLILSGTP IQNNVLELWS LFDFLMPGFL GTEKQFVQRF SRPILSSRDA1560
1561 KSSAKEQEAG VLAMEALHRQ VLPFLLRRVK EDVLKDLPPK ITQDLLCELS PLQLRLYEDF1620
1621 SNKHLKDCLD KLGDSSSASM VTENLSAKTH IFQALRYLQN VCNHPKLVLR QSEELTKVTS1680
1681 QLALSNSSLD DIEHSAKLPA LKQLLLDCGI GVQTESVSQH RALIFCQLKA MLDIVEQDLL1740
1741 RRHLPSVTYL RLDGSVPASQ RQDIVNNFNS DPSIDVLLLT TMVGGLGLNL TGADTVIFVE1800
1801 HDWNPMKDLQ AMDRAHRIGQ KKVVNVYRLI TRNSLEEKIM GLQKFKILTA NTVVSAENAS1860
1861 LQTMGTSQIF DLFNGGKDKG AESGSSAVQG TASGGMSMNT IIENLPELWS EHQYEEEYDL1920
1921 GNFVQALKK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
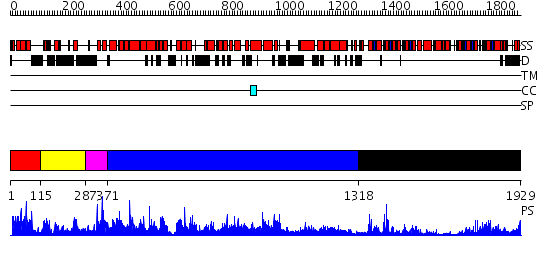
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..114] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [115..286] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [287..370] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [371..1317] | 2.48 | No description for 2ie3A was found. | |
| 5 | View Details | [1318..1929] | 97.522879 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.45 |
Source: Reynolds et al. (2008)