













| General Information: |
|
| Name(s) found: |
CG5205-PA /
FBpp0082601
[FlyBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2183 amino acids |
Gene Ontology: |
|
| Cellular Component: |
U5 snRNP
[ISS]
small nuclear ribonucleoprotein complex [ISS] |
| Biological Process: |
nuclear mRNA splicing, via spliceosome
[ISS]
|
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] RNA helicase activity [ISS] ATP-dependent helicase activity [IEA] helicase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWEPPRLSAS LRAKTELEAR SRRLNVLRQA RTTSAASTTT NSQNKEEQTI ARLLAQMPTE 60
61 KQPAAKVQLQ KLRNLVQECM GYEEQPQVVE HAALFLFWLL LDQRVLLVAT TQRLHGMFGQ 120
121 TFDRKKTQIH DCVQAIGDLL EDHERSALKR WRQTHHKVPG VGPLWGAHIH VKCTPSEWMD 180
181 ISLLTDLAPD ALLKNRTVNK FSMKHTTKAA PVASTSSEAA KLSPELQTLL EISAKLDDEH 240
241 LIIRVGDILG SQRSSEVLQN ELMEILGFDY FELVEKLLME RDKIARQLDQ FATRSRRVME 300
301 VKQKRMEAAA SGGAAERRPT VASAVVVQSA QEKQLSKMQR REEKKLQRIM RSIKDEEAED 360
361 DPNCAVAVSV QQLRMQHQRK LLEAAQREPL LLSTKAAKAE YKQSSYNQPI HYPYVFDSQL 420
421 LAKQHAGFIG GSRITLPDNA QRIDNKQWEE VKIPASEPPP LSVGNKRVQI EELDDVGRLA 480
481 FANCKELNRI QSVVFPVAYH SNENMLVCAP TGAGKTNVAM LSIVHTIRCH LEQGVINRDE 540
541 FKIVYIAPMK ALAAEMVDNF SKRLKSLQIA VRELTGDIQL TKAEMAATQI LVTTPEKWDV 600
601 VTRKGSGDVA LISLVELLII DEVHLLHGER GPVVEALVAR TLRLVESSQS MIRIVGLSAT 660
661 LPNYIDVAHF LRVNPMKGLF YFDSRFRPVP LDTNFVGIKS VKQLQQIADM DQCCYQKCVE 720
721 MVQEGHQIMV FVHARNATVR TANVIRELAQ QNNTSALFLP KDSAAHGLAT RSIQRSRNKQ 780
781 LVELFSCGLA MHHAGMLRAD RQMVEKYFVE GHISVLVCTA TLAWGVNLPA HAVIIRGTDI 840
841 YDAKHGSFVD LGILDVLQIF GRAGRPQFDK SGVGTIITSY DKLNHYLSLL TNQFPIESNF 900
901 VNCLADNLNA EIGLGTITNV DEAIEWLSYT YLFVRMRINP HVYGIEYSEL EKDPTLEARR 960
961 RALIMSAAMS LDKARMMRFN QRTMDMNITD LGRTASYFYI KYDTVETFNE LMKPFMTQAE1020
1021 ILAMISQAQE FQQLKVRDDE MEELDELKNA YCKIKPYGGS ENVHGKVNIL IQTYLSNGYV1080
1081 KSFSLSSDMS YITTNIGRIS RALFSIVLRQ NNAVLSGNML QLCKMFERRQ WDFDCHLKQF1140
1141 PTINAETIDK LERRGLSVYR LRDMEHRELK EWLRSNTYAD LVIRSAHELP LLEVEASLQP1200
1201 ITRTVLRIKV DIWPSFTWND RVHGKTCQSF WLWIEDPESN YIYHSELFQV TRKLVMSGQS1260
1261 QQLVMTIPLK EPLPPQYYIR VSSDNWLGST TCIPLSFQHL VLPEHHPPLT ELLPLRPLPV1320
1321 SCLKNVVYES LYKFTHFNPI QTQIFHCLYH TDNNVLLGAP TGSGKTIVAE IAIFRALNQN1380
1381 PKCKVVYIAP LKALVKERIA DWEQRFQRSS LGLKVVELTG DVTPDIQAIR ESQLIVTTPE1440
1441 KWDGISRSWQ TREYVQHVSL IVIDEIHLLG EDRGPVIEVI VSRTNFISSH TGRAIRIVGL1500
1501 STALANAQDL ANWLGINKMG LYNFKPSVRP VPLQVHINGF PGKHYCPRMA TMNRPTFQAI1560
1561 RTYSPCEPTI VFVSSRRQTR LTALDLITFV AGESNPKQFL HIPEDEIELI LQNIREQNLK1620
1621 FCLAFGIGLH HAGLQEQDRK CVEELFLNRK IQILVATATL AWGVNLPAHL VVIKGTEYFD1680
1681 GKVKKYVDMP ITDVLQMMGR AGRPQFDNEG VAVVLVHDEK KNFYKKFLYD PFPVESSLLG1740
1741 VLPEHINAEI VAGTVQSKQA ALDYLTWTYF FRRLLRNPSY YQLQDIEPEN VNNFMSNLVE1800
1801 RVVYELSAAA CLVERDGCLI PTFLGRISSY YYLSYRTMKH FLEDLQPGMN TKKVLLAIAD1860
1861 SYEFDQLPVR HNEDKHNEEM AEVSRFRPPS SSWDSSYTKT FLLLQAHFAR QSLPNSDYLT1920
1921 DTKSALDNAT RVMQAMVDYT AERGWLSTTL VVQQLMQSVI QARWFDGSEF LTLPGVNEDN1980
1981 LDAFLNIPHD DYDYLTLPVL KELCKQEYEV LAKPLRDAFE EHEIEKMYKV IQDLPEIAIQ2040
2041 IFVEGRHMEN EYAKRPLSLS DDTRGEWMSL HANEDYVLIV NLQRLNVSGQ RRGGGQSYTV2100
2101 HCPKYPKPKN EAWFLTLGSQ ANDELLAMKR VSIRGQRCTN RISFQATPRL GRLQLTLYLM2160
2161 SDCLMGFDQQ YDLQFEIIDA KEV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
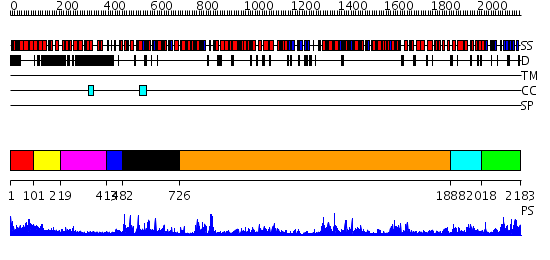
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..100] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [101..218] | 1.139989 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [219..412] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [413..481] | 71.30103 | No description for 2j0qA was found. | |
| 5 | View Details | [482..725] | 6.522879 | No description for 2v8oA was found. | |
| 6 | View Details | [726..1887] | 47.69897 | Crystal structure of Escherichia coli transcription-repair coupling factor | |
| 7 | View Details | [1888..2017] | 84.0 | No description for 2p6rA was found. | |
| 8 | View Details | [2018..2183] | 79.69897 | No description for 2q0zX was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)