













| General Information: |
|
| Name(s) found: |
Su(fu)-PA /
FBpp0082134
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 468 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
protein complex [IPI] Hedgehog signaling complex [IPI] |
| Biological Process: |
negative regulation of transcription factor activity
[IGI]
cytoplasmic sequestering of transcription factor [IMP] negative regulation of smoothened signaling pathway [IGI][NAS] |
| Molecular Function: |
transcription factor binding
[IPI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEANLDKKP EVKPPPGLKA IIDHLGQVYP NQPNPLQVTT LLKYWLGGQD PLDYISMYNY 60
61 PGDVDRNVPP HWHYISFGLS DLHGDERVHL REEGVTRSGM GFELTFRLAK TEIELKQQIE 120
121 NPEKPQRPPT WPANLLQAIG RYCFQTGNGL CFGDNIPWRK SLDGSTTSKL QNLLVAQDPQ 180
181 LGCIDTPTGT VDFCQIVGVF DDELEQASRW NGRGVLNFLR QDMQTGGDWL VTNMDRQMSV 240
241 FELFPETLLN LQDDLEKQGS DLAGVNADFT FRELKPTKEV KEEVDFQALS EKCANDENNR 300
301 QLTDTQMKRE EPSFPQSMSM SSNSLHKSCP LDFQAQAPNC ISLDGIEITL APGVAKYLLL 360
361 AIKDRIRHGR HFTFKAQHLA LTLVAESVTG SAVTVNEPYG VLGYWIQVLI PDELVPRLME 420
421 DFRSAGLDEK CEPKERLELE WPDKNLKLII DQPEPVLPMS LDAAPLKM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
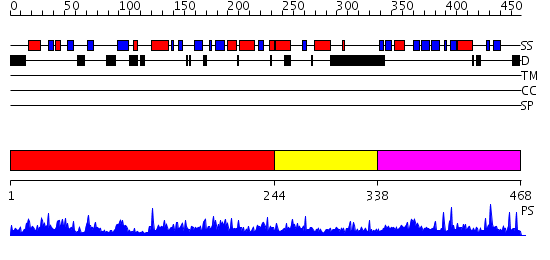
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..243] | 118.0 | Human Suppressor of Fused (N-terminal domain) | |
| 2 | View Details | [244..337] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [338..468] | 1.01197 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)