













| General Information: |
|
| Name(s) found: |
grn-PA /
FBpp0081304
[FlyBase]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 486 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS][NAS]
|
| Biological Process: |
regulation of transcription, DNA-dependent
[IEA]
tissue development [IMP][NAS] regulation of transcription from RNA polymerase II promoter [ISS] organ morphogenesis [IMP][NAS] |
| Molecular Function: |
zinc ion binding
[IEA]
transcription factor activity [IEA] RNA polymerase II transcription factor activity [NAS] general RNA polymerase II transcription factor activity [ISS][NAS] sequence-specific DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDMTSTAEAA ARSWYDSPRL GGGGSSGGGN GGGVSPQTNG LGSAGSSLAH SHHSLSSGAS 60
61 SAGSSVGVGS ALGGGGGSGL DTSDMSAFYA LESNGHHRRY YPSYHQHTSR MPSTHASPQV 120
121 CRPHFHTPLS PWLTSEHKSF APASAWSMGQ FACPQEPQVE HKLGQMGQSH QTTAAGQHSF 180
181 PFPPTPPKDS TPDSVQTGPS EYQAVMNAFM HQQATGSTSL TDASCALDIK PSIQNGSASG 240
241 SSGSGTTHTS TPKQREEGRE CVNCGATSTP LWRRDGTGHY LCNACGLYYK MNGQNRPLIK 300
301 PKRRLTLQSL QSAAKRAGTS CANCKTTTTT LWRRNASGEP VCNACGLYYK LHNVNRPLTM 360
361 KKEGIQTRNR KLSSKSKKKK GLGGGCLPIG GHLGMGDFKP LDPSKGFGGG FSASMAQHGH 420
421 LSSGLHPAHA HMHGSWYTGG MGALGASSGL QGGFSTAGSL SGAVVPHSQP YHLGLSSMGT 480
481 WRTDYT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
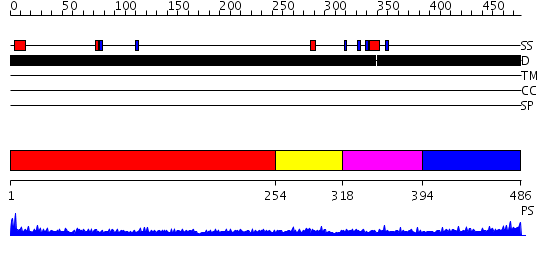
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..253] | 3.211995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [254..317] | 2.95 | Erythroid transcription factor GATA-1 | |
| 3 | View Details | [318..393] | 10.69897 | Erythroid transcription factor GATA-1 | |
| 4 | View Details | [394..486] | 2.17899 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)