













| General Information: |
|
| Name(s) found: |
FBpp0309997
[FlyBase]
enok-PA / FBpp0072159 [FlyBase] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 2291 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleosome
[IEA]
histone acetyltransferase complex [ISS] |
| Biological Process: |
neuroblast proliferation
[IMP]
mushroom body development [IMP] regulation of transcription [IEA] nucleosome assembly [IEA] |
| Molecular Function: |
histone acetyltransferase activity
[ISS]
DNA binding [IEA] zinc ion binding [IEA] protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMRESAHDIN MDTWNQWILE AISKIRSQKQ RPSVQRICQA IGTHHKFHED IVAEKLEKAV 60
61 ESGAVIKVYN KGLHSYKAPM AKRRVKVDKN TNLYKLVAKA VHDLGECEGS TIKSIENYIQ 120
121 KFNCIDLSPN VDFKAVIKAS IKKAVDAGFL IQEGKLYKKG KSLTTPRKCA PVSEVVIKGE 180
181 ESCTHCSGNS QKNLNGIPEP LSSCKQCGIS LHTTCANIAG RCKSQSYVLL YMLVTKGTIW 240
241 DCQNCADCAV CKMRNRGPCL LQCFVCKDHF HLTCLDTIPD KKPKHPYRCK TCSKQGFDTP 300
301 KLVKKDNSRI KMEAERMTPS YKYASNKRQS IKKDGNIESP STSKLCVYND GNGQRRKVPA 360
361 SLSSRKMNHS EDQFEFPSKQ KRSQILQGGY LASQETRKRT FSDLSSTSSS SESEDEDDED 420
421 DDRNRNGDDN DQDESTSSDS CTSSSSDSDS SESSSDTSEW DDEYDEEEDY DTDRSTIREK 480
481 NVFESGKQSC AKLSTGDGLG SPQNNELGSE NWGFAAVAKN PIDIFVKSKH NSNVKGIGYS 540
541 KPAASTFATS PAANRVQKST PVASSTPEKN SAPIGGMTIK RQTANGQKKK IVPLKSMPLE 600
601 AGKSYEKCED DIPYLTEETV MKVQLLEEIE RSRETHSQAD GKREMNNDED APGESTKRAN 660
661 PFENQPLPPG VTATDVELYQ EVLHKAVVQM SNNHIEKVND VSLKSGQNTQ SPKSIQIGKW 720
721 DIETWYSSPF PQEYARLLKL FLCEFCLKYT KSRSVLDRHQ NKCIWKQPPG TEIFRQGNIS 780
781 VFEVDGNVNK IYCQNLCLLA KFFLDHKTLY YDVEPFLFYI LTKNDQSGCH LVGYFSKEKH 840
841 CTQKYNVSCI LTMPQYQRQG YGRFLIDFSY LLSREEGQLG TPEKPLSDLG RLSYFSYWKS 900
901 VVLEYLYKHR NYTKITFKDI AIKTGLAISD IALAFELLNF IKLRKNDGDI RYQINVKIEW 960
961 KKVLAHHNKM ANSKTRIIIE PDCLRWSPLL SVSKLPNIIK PISNREYLSN QMETNSSFKE1020
1021 INEDKISVKQ GVSTSSKVNQ ESSELSTKNR GPSSEKLIDE RENQRKRAYP EEFLKSASNE1080
1081 AKKPKLTAPQ PNLEDQSFGE LSDSSDCKSS KDLSEQLTKR AQRLAKRNDL IPHTNKRKHF1140
1141 DRPLENNTTS TDHNPPAQSQ ADTAGTENGH KSEETAAKEN CKNVVEKEHV DKPVNIMPKL1200
1201 RGKQSNGKRS EEPQFNENPT SDPVAVASLS PAKEITREPV KVAKTESPGK NQESAKHDAE1260
1261 PNSVSDQVLD AVAKKTKKTL FSDVASYNTE DVKYNSEEKP MAIAQEPKES PTEAGIKETK1320
1321 PLEVPEQKAK AKSPEKPATV AETVLQHVHA PVNTEKISTE ESKLELDVHY LKLSPDSALN1380
1381 PPVASVQKPV PEPPAPKEKA KDLQAAQPSE ALQKQPEIEM VKKTETKTET PVSNPPSNRT1440
1441 DVSSLVEQPV KAAPEKVEQP MVKEAVDIKD KALKKPVPDV PVVKPEATSC EKKIDHSKTK1500
1501 VFHEKEAIKT DQQLVQVKEE EKSNARAPSA PIPIRDKIQL KGNEHAQLQN SIPSQFPLNQ1560
1561 MPNYHTSQYW QWEYYGYNLS HLDASQKGQK QFHKDLATTM AYTHNFTQNL YQSANLAMQH1620
1621 AHHQMHQTKE KHKVERKNSG KKEEQNKVVS SVNVVSTARE DAHVQHCNEY AANNQAALYN1680
1681 QKCASQQKQA QQMKSLANVQ NPVPRQSNSN APESTVLMPN PVISQAGSVP AKQKVEHGVN1740
1741 STSVISREGK LNKIGHSNIH ATAQHANLQV GGGQSDVNPN MVYNTESSAN SAMQHYDCGI1800
1801 SAQINMDSPA NIGSAIAHGP QEISATSNVH MHQPHRQFSD CSMQNQPVTT PMHMSIQNSH1860
1861 MQQQNNLNLN LAPEGSSNIN LLSNPQHQHQ SRKLNAQADI AVSSPTPSHR ATTPKQIRSG1920
1921 NTQQRDAKNA APPTTTTNTH HQIPQGGAAA NISKSHHDSL HNLQFSQHGH QQNMQPIDYV1980
1981 PIPQISQNFS ANPSNYDIVG MPAVIQQRMS LNGSVHSLAN SHQRIEQPSS ACAVNNFYLQ2040
2041 NNMPAAENAP RVPVSSSLGG PPTAGNNDQR QAGQDSITTA NSSGATASLA GNLCSLSKLQ2100
2101 QLTNCLESQP CNTSPGAQVN LAPSPHHPIP PNSTPPPHLL MQNRNISTPP NMLQTQVTPL2160
2161 QYKYYPGNMN IPPITSAQNT SRNTRNTPSA PVQHTSTPMG SGNNRTANVH ISPNLMAPYG2220
2221 AINSYRMSPQ QSPPTGSYSS GGDYPNSQIP MQMMNMQSQY QDACVLQRGT PSNPMYPTYS2280
2281 PYLPLNGSIR R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
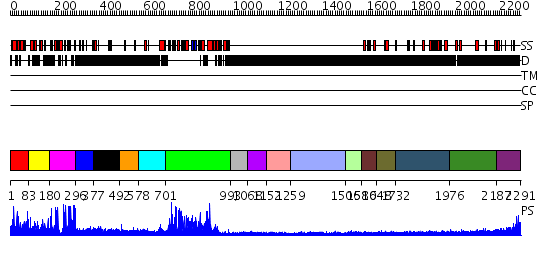
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [83..179] | 3.78 | YEAST HISTONE H1 GLOBULAR DOMAIN II, HHO1P GII, SOLUTION NMR STRUCTURES | |
| 3 | View Details | [180..295] | 25.39794 | No description for 2ysmA was found. | |
| 4 | View Details | [296..376] | 7.30103 | No description for 2f6jA was found. | |
| 5 | View Details | [377..491] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [492..577] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [578..700] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [701..992] | 134.0 | No description for 2ozuA was found. | |
| 9 | View Details | [993..1067] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1068..1151] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1152..1258] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1259..1505] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [1506..1579] | N/A | No confident structure predictions are available. | |
| 14 | View Details | [1580..1647] | N/A | No confident structure predictions are available. | |
| 15 | View Details | [1648..1731] | N/A | No confident structure predictions are available. | |
| 16 | View Details | [1732..1975] | N/A | No confident structure predictions are available. | |
| 17 | View Details | [1976..2186] | N/A | No confident structure predictions are available. | |
| 18 | View Details | [2187..2291] | 2.347987 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 13 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 14 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 15 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 16 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 17 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 18 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)