













| General Information: |
|
| Name(s) found: |
FBpp0300559
[FlyBase]
synj-PA / FBpp0071671 [FlyBase] synj-PB / FBpp0071670 [FlyBase] |
| Description(s) found:
Found 42 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1218 amino acids |
Gene Ontology: |
|
| Cellular Component: |
terminal button
[IDA]
vesicle membrane [IDA] |
| Biological Process: |
maintenance of presynaptic active zone structure
[IMP]
synaptic vesicle endocytosis [IMP][TAS] regulation of synapse structure and activity [IMP] dephosphorylation [IEA][NAS] neurotransmitter secretion [NAS] |
| Molecular Function: |
inositol trisphosphate phosphatase activity
[NAS]
RNA binding [IEA] phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity [IEA] inositol-polyphosphate 5-phosphatase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMSKVIRVL EKSIAPSPHS VLLEHRNKSD SILFESHAVA LLTQQETDVI RKQYTKVCDA 60
61 YGCLGALQLN AGESTVLFLV LVTGCVSMGK IGDIEIFRIT QTTFVSLQNA APNEDKISEV 120
121 RKLLNSGTFY FAHTNASASA SGASSYRFDI TLCAQRRQQT QETDNRFFWN RMMHIHLMRF 180
181 GIDCQSWLLQ AMCGSVEVRT VYIGAKQARA AIISRLSCER AGTRFNVRGT NDEGYVANFV 240
241 ETEQVIYVDG DVTSYVQTRG SVPLFWEQPG VQVGSHKVKL SRGFETSAAA FDRHMSMMRQ 300
301 RYGYQTVVNL LGSSLVGSKE GEAMLSNEFQ RHHGMSAHKD VPHVVFDYHQ ECRGGNFSAL 360
361 AKLKERIVAC GANYGVFHAS NGQVLREQFG VVRTNCLDCL DRTNCVQTYL GLDTLGIQLE 420
421 ALKMGGKQQN ISRFEEIFRQ MWINNGNEVS KIYAGTGAIQ GGSKLMDGAR SAARTIQNNL 480
481 LDNSKQEAID VLLVGSTLSS ELADRARILL PSNMLHAPTT VLRELCKRYT EYVRPRMARV 540
541 AVGTYNVNGG KHFRSIVFKD SLADWLLDCH ALARSKALVD VNNPSENVDH PVDIYAIGFE 600
601 EIVDLNASNI MAASTDNAKL WAEELQKTIS RDNDYVLLTY QQLVGVCLYI YIRPEHAPHI 660
661 RDVAIDCVKT GLGGATGNKG ACAIRFVLHG TSMCFVCAHF AAGQSQVAER NADYAEITRK 720
721 LAFPMGRTLK SHDWVFWCGD FNYRIDMEKD ELKECVRNGD LSTVLEFDQL RKEQEAGNVF 780
781 GEFLEGEITF DPTYKYDLFS DDYDTSEKQR APAWTDRVLW RRRKALAEGD FAASAWNPGK 840
841 LIHYGRSELK QSDHRPVIAI IDAEIMEIDQ QRRRAVFEQV IRDLGPPDST IVVHVLESSA 900
901 TGDEDGPTIY DENVMSALIT ELSKMGEVTL VRYVEDTMWV TFRDGESALN ASSKKSIQVC 960
961 GLDLILELKS KDWQHLVDSE IELCTTNTIP LCANPVEHAQ LLQAITPELP QRPKQPPTRP1020
1021 PARPPMPMSP KNSPRHLPHV GVISIVPKPA KPPMPPQPQS QPLIPSPLQP QVAPPRPPAM1080
1081 DTTPSSKSQS PTELVSASSS TSSSGKTSPT APAVLSGPPT PPRQMQSKQA TPVSTPTRQV1140
1141 GSPKDPIPSD TAYETASNIY EEIQEDVPAP RHPPPAGPPP VIGAPMGPPP PLPNRRGPPP1200
1201 IPNRSGNAPP LPTRPANN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
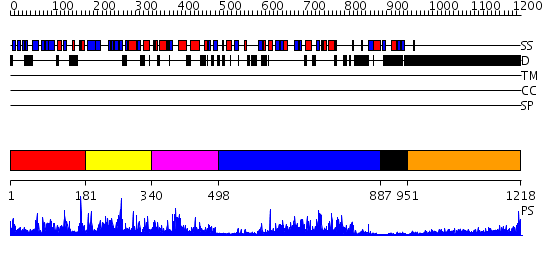
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..180] | 2.181973 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [181..339] | 1.193949 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [340..497] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [498..886] | 87.045757 | Synaptojanin, IPP5C domain | |
| 5 | View Details | [887..950] | 2.86 | Solution structure of RNP domain in Synaptojanin 2 | |
| 6 | View Details | [951..1218] | 7.69897 | No description for 1y0fA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)