













| General Information: |
|
| Name(s) found: |
opa1-like-PB /
FBpp0086699
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 933 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
pupariation
[IMP]
mitochondrial fusion [IMP] |
| Molecular Function: |
GTP binding
[ISS]
GTPase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRIYQNTYR RTARKAVVYS TKVACCNHST LCGITSHPRR AQDSGSSSSN GRHRGHEEFL 60
61 LAGNPARGWQ MPPPSRGYGM LVVRILRGAL KLRYIVLGGA IGGGVSLSKK YEEWKDGLPN 120
121 FKWLEDAMPQ GERWSQFSRN LIEVGSLVKN AIEVAKDDLK AKTTVAALGI TSDESRKKYE 180
181 KLQSQVETLQ TEIMNVQIKY QKELEKMEKE NRELRQQYLI LKTNKKTTAK KIKKSLIDMY 240
241 SEVLDELSGY DTGYTMADHL PRVVVVGDQS SGKTSVLESI AKARIFPRGS GEMMTRAPVK 300
301 VTLAEGPYHV AQFRDSDREY DLTKESDLQD LRRDVEFRMK ASVRGGKTVS NEVIAMTVKG 360
361 PGLQRMVLVD LPGIISTMTV DMASDTKDSI HQMTKHYMSN PNAIILCIQD GSVDAERSNV 420
421 TDLVMQCDPL GRRTIFVLTK VDLAEELADP DRIRKILSGK LFPMKALGYY AVVTGRGRKD 480
481 DSIDAIRQYE EDFFKNSKLF HRRGVIMPHQ VTSRNLSLAV SDRFWKMVRE TIEQQADAFK 540
541 ATRFNLETEW KNNFPRLRES GRDELFDKAK GEILDEVVTL SQISAKKWDD ALSTKLWEKL 600
601 SNYVFESIYL PAAQSGSQNS FNTMVDIKLR QWAEQALPAK SVEAGWEALQ QEFISLMERS 660
661 KKAQDHDGIF DQLKSAVVDE AIRRHSWEDK AIDMLRVIQL NTLEDRFVHD KQEWDSAVKF 720
721 LESSVNAKLV QTEETLAQMF GPGQMRRITH WQYLTQDQQK RRSVKNELDK ILKNDTKHLP 780
781 TLTHDELTTV RKNLQRDNVD VDTDYIRQTW FPVYRKHFLQ QALQRAKDCR KAYYLYTQQG 840
841 AECEISCSDV VLFWRIQQVI KITGNALRQQ VINREARRLD KEIKAVLDEF SDDEEKKGYL 900
901 LTGKRVLLAE ELIKVRQIQE KLEEFINSLN QEK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
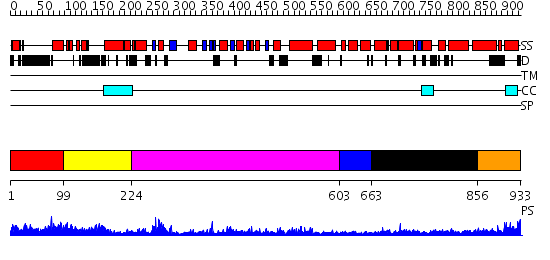
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [99..223] | 30.522879 | Structure of the GTP-binding protein TrmE from Thermotoga maritima | |
| 3 | View Details | [224..602] | 64.0 | Structure of the nucleotide-free myosin II motor domain from Dictyostelium discoideum fused to the GTPase domain of dynamin 1 from Rattus norvegicus | |
| 4 | View Details | [603..662] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [663..855] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [856..933] | 2.32799 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)