













| General Information: |
|
| Name(s) found: |
alh-6 /
CE40074
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 423 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IEA |
| Biological Process: |
cellular aldehyde metabolic process
[IEA locomotory behavior [IMP proline metabolic process [IEA negative regulation of transcription [IEA positive regulation of growth rate [IMP metabolic process [IEA proline biosynthetic process [IEA gamete generation [IMP valine metabolic process [IEA betaine biosynthetic process [IEA positive regulation of locomotion [IMP |
| Molecular Function: |
1-pyrroline-5-carboxylate dehydrogenase activity
[IEA DNA binding [IEA betaine-aldehyde dehydrogenase activity [IEA proline dehydrogenase activity [IEA oxidoreductase activity [IEA aldehyde dehydrogenase [NAD(P)+] activity [IEA methylmalonate-semialdehyde dehydrogenase (acylating) activity [IEA transcription repressor activity [IEA oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLNASTMLG QGKNIIQAEI DAACELIDFF RFNAVFAMEL EHYEPISTKI TKNTMQFRGM 60
61 EGFVAAIAPF NFTAIGGNLP TAPALMGNVS LWKPSNTAVL SNYIIYELLE EAGMPPGILS 120
121 FLPSDGPVFG DVITASPHLS AVNFTGSVPT FKTIWRKVAE NLDNYVTFPK LIGECGGKNF 180
181 HFVHPSAHVD AVAAGTARSA WEYSGQKCSA CSRMYAPKSI WPKILEKISA IHKEIKLGDV 240
241 RDGSVFLSAV IDDKAFARLK AYIDFAKTGA DGANVVLGGK CDDKTGYFIE PTLITVTDPK 300
301 SKLLTEEMFG PVVTVLVYED SKVDEVLATV KDATPYGLTG AVFSQDKEFL YRARDVLRDA 360
361 VGNMYLNDKS TGSIVGQQPF GGSRLSGTND KAGGPHYGLR WTSPLTIKET SVSLTDWKYP 420
421 SME |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
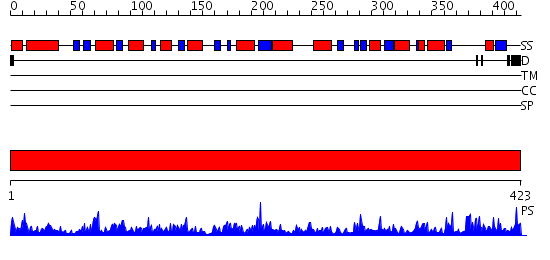
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..423] | 102.0 | 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE |
Functions predicted (by domain):
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)