













| General Information: |
|
| Name(s) found: |
FKH2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 642 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVRRLESKS EHISDDEERK EQLDYKKQMD VDTDRNIVLN GRLESQIAKL SVPPHEMRVV 60
61 DDYSNSKNAE RHSGEIQAYA KFAGSTWTYY VKKIRIILGR EPANPSPKGK NEDLEVIDMN 120
121 FGPSKVVSRK HAVVEYDLDD QTWNCSVYGR NGIKVDGKLF KNGETVKLTS GSILEVAGLQ 180
181 MMFVLPNAAE QKQTDESTIK EDAIKSEISA AVNDAAEYGD NKKPPYSYSV MIAQAILSSS 240
241 ECMMTLSNIY SWISTHYPYY RTTKSGWQNS IRHNLSLNKA FRKVPRKSGE QGKGMKWSIV 300
301 PEFREEFIAK TRKTPRKRSP SSPVPLLAKK REGSPSLPIP ILPKMKDTSI PAAEPASSTT 360
361 SARDQTPSTP KDVGSPSTAE TSAEEKQMET YKTPTHAALS DIISTHDYAL DANSASQTKK 420
421 AAFGSPIGSS TYPTSSPAPF WKYVAVPNPH DWPQVGSYDT ISPYRNPVNS HLIYSQIQQS 480
481 SPKKIDEQLH DLQGVDLVNG FEGISSWRES MVNKLRSSVS DSPTMNLANS NSKSSPVAVQ 540
541 RVSTLPQASA NKQAKEMESK MSNSPTQKSK TEENNQAVRA ILDASATMEK QYDLHRLPTP 600
601 TSQTESASVP QIANPPNSQN LVKEKSPQQY IQVPQSNVKS SA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
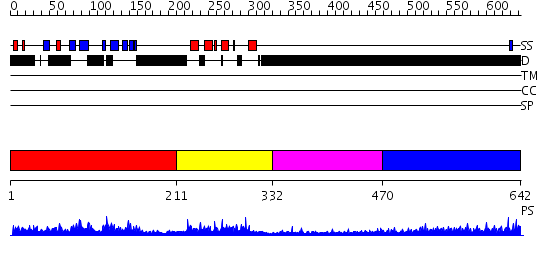
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..210] | 4.31 | Phosphotyrosine binding domain of Rad53 | |
| 2 | View Details | [211..331] | 41.522879 | Adipocyte-transcription factor FREAC-11 (s12, fkh-14) | |
| 3 | View Details | [332..469] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [470..642] | 9.308997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)