













| General Information: |
|
| Name(s) found: |
SMAD2
[HGNC (HUGO)]
|
| Description(s) found:
Found 55 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 467 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSILPFTPP VVKRLLGWKK SAGGSGGAGG GEQNGQEEKW CEKAVKSLVK KLKKTGRLDE 60
61 LEKAITTQNC NTKCVTIPST CSEIWGLSTP NTIDQWDTTG LYSFSEQTRS LDGRLQVSHR 120
121 KGLPHVIYCR LWRWPDLHSH HELKAIENCE YAFNLKKDEV CVNPYHYQRV ETPVLPPVLV 180
181 PRHTEILTEL PPLDDYTHSI PENTNFPAGI EPQSNYIPET PPPGYISEDG ETSDQQLNQS 240
241 MDTGSPAELS PTTLSPVNHS LDLQPVTYSE PAFWCSIAYY ELNQRVGETF HASQPSLTVD 300
301 GFTDPSNSER FCLGLLSNVN RNATVEMTRR HIGRGVRLYY IGGEVFAECL SDSAIFVQSP 360
361 NCNQRYGWHP ATVCKIPPGC NLKIFNNQEF AALLAQSVNQ GFEAVYQLTR MCTIRMSFVK 420
421 GWGAEYRRQT VTSTPCWIEL HLNGPLQWLD KVLTQMGSPS VRCSSMS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
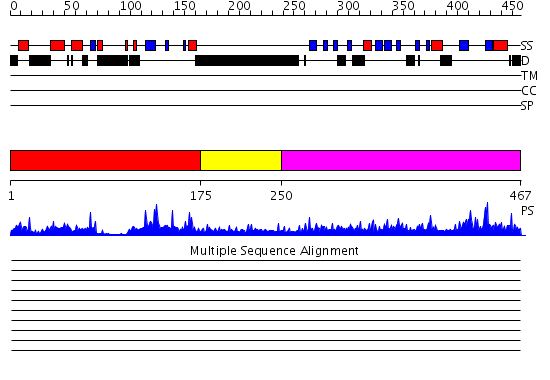
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..174] | 57.30103 | Crystal structure of Smad3-MH1 bound to DNA at 2.4 A resolution | |
| 2 | View Details | [175..249] | 2.089996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [250..467] | 106.0 | Smad2 MH2 domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)