













| General Information: |
|
| Name(s) found: |
AMFR
[HGNC (HUGO)]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 643 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLLFLERFP WPSLRTYTGL SGLALLGTII SAYRALSQPE AGPGEPDQLT ASLQPEPPAP 60
61 ARPSAGGPRA RDVAQYLLSD SLFVWVLVNT ACCVLMLVAK LIQCIVFGPL RVSERQHLKD 120
121 KFWNFIFYKF IFIFGVLNVQ TVEEVVMWCL WFAGLVFLHL MVQLCKDRFE YLSFSPTTPM 180
181 SSHGRVLSLL VAMLLSCCGL AAVCSITGYT HGMHTLAFMA AESLLVTVRT AHVILRYVIH 240
241 LWDLNHEGTW EGKGTYVYYT DFVMELTLLS LDLMHHIHML LFGNIWLSMA SLVIFMQLRY 300
301 LFHEVQRRIR RHKNYLRVVG NMEARFAVAT PEELAVNNDD CAICWDSMQA ARKLPCGHLF 360
361 HNSCLRSWLE QDTSCPTCRM SLNIADNNRV REEHQGENLD ENLVPVAAAE GRPRLNQHNH 420
421 FFHFDGSRIA SWLPSFSVEV MHTTNILGIT QASNSQLNAM AHQIQEMFPQ VPYHLVLQDL 480
481 QLTRSVEITT DNILEGRIQV PFPTQRSDSI RPALNSPVER PSSDQEEGET SAQTERVPLD 540
541 LSPRLEETLD FGEVEVEPSE VEDFEARGSR FSKSADERQR MLVQRKDELL QQARKRFLNK 600
601 SSEDDAASES FLPSEGASSD PVTLRRRMLA AAAERRLQKQ QTS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
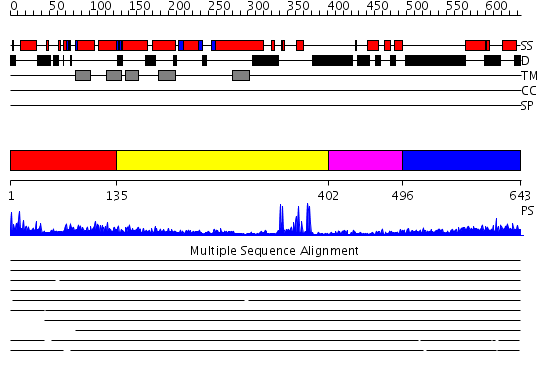
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [135..401] | 24.39794 | Cbl; N-terminal domain of cbl (N-cbl); CBL | |
| 3 | View Details | [402..495] | 3.15 | No description for 2ekfA was found. | |
| 4 | View Details | [496..643] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|