













| General Information: |
|
| Name(s) found: |
SLC25A13
[HGNC (HUGO)]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 675 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAAKVALTK RADPAELRTI FLKYASIEKN GEFFMSPNDF VTRYLNIFGE SQPNPKTVEL 60
61 LSGVVDQTKD GLISFQEFVA FESVLCAPDA LFMVAFQLFD KAGKGEVTFE DVKQVFGQTT 120
121 IHQHIPFNWD SEFVQLHFGK ERKRHLTYAE FTQFLLEIQL EHAKQAFVQR DNARTGRVTA 180
181 IDFRDIMVTI RPHVLTPFVE ECLVAAAGGT TSHQVSFSYF NGFNSLLNNM ELIRKIYSTL 240
241 AGTRKDVEVT KEEFVLAAQK FGQVTPMEVD ILFQLADLYE PRGRMTLADI ERIAPLEEGT 300
301 LPFNLAEAQR QKASGDSARP VLLQVAESAY RFGLGSVAGA VGATAVYPID LVKTRMQNQR 360
361 STGSFVGELM YKNSFDCFKK VLRYEGFFGL YRGLLPQLLG VAPEKAIKLT VNDFVRDKFM 420
421 HKDGSVPLAA EILAGGCAGG SQVIFTNPLE IVKIRLQVAG EITTGPRVSA LSVVRDLGFF 480
481 GIYKGAKACF LRDIPFSAIY FPCYAHVKAS FANEDGQVSP GSLLLAGAIA GMPAASLVTP 540
541 ADVIKTRLQV AARAGQTTYS GVIDCFRKIL REEGPKALWK GAGARVFRSS PQFGVTLLTY 600
601 ELLQRWFYID FGGVKPMGSE PVPKSRINLP APNPDHVGGY KLAVATFAGI ENKFGLYLPL 660
661 FKPSVSTSKA IGGGP |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample pIC146 from april 2005 | Okada, M., et al. (2006) | |
| View Run | No Comments | Cheeseman, I.M., et al. (2004) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
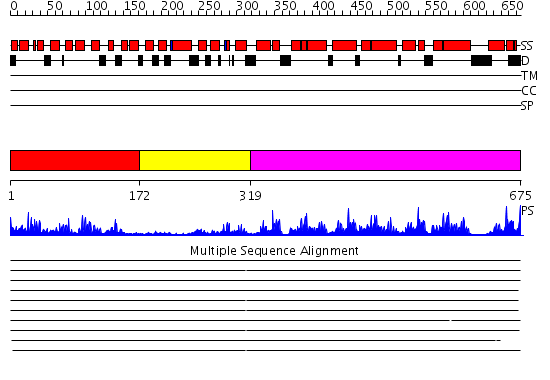
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 45.30103 | No description for 1zuzA was found. | |
| 2 | View Details | [172..318] | 4.52 | Structure of calmodulin bound to a calcineurin peptide: a new way of making an old binding mode | |
| 3 | View Details | [319..675] | 83.221849 | structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.67 |
Source: Reynolds et al. (2008)