













| General Information: |
|
| Name(s) found: |
MCM6
[HGNC (HUGO)]
|
| Description(s) found:
Found 38 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 821 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLAAAAEPG AGSQHLEVRD EVAEKCQKLF LDFLEEFQSS DGEIKYLQLA EELIRPERNT 60
61 LVVSFVDLEQ FNQQLSTTIQ EEFYRVYPYL CRALKTFVKD RKEIPLAKDF YVAFQDLPTR 120
121 HKIRELTSSR IGLLTRISGQ VVRTHPVHPE LVSGTFLCLD CQTVIRDVEQ QFKYTQPNIC 180
181 RNPVCANRRR FLLDTNKSRF VDFQKVRIQE TQAELPRGSI PRSLEVILRA EAVESAQAGD 240
241 KCDFTGTLIV VPDVSKLSTP GARAETNSRV SGVDGYETEG IRGLRALGVR DLSYRLVFLA 300
301 CCVAPTNPRF GGKELRDEEQ TAESIKNQMT VKEWEKVFEM SQDKNLYHNL CTSLFPTIHG 360
361 NDEVKRGVLL MLFGGVPKTT GEGTSLRGDI NVCIVGDPST AKSQFLKHVE EFSPRAVYTS 420
421 GKASSAAGLT AAVVRDEESH EFVIEAGALM LADNGVCCID EFDKMDVRDQ VAIHEAMEQQ 480
481 TISITKAGVK ATLNARTSIL AAANPISGHY DRSKSLKQNI NLSAPIMSRF DLFFILVDEC 540
541 NEVTDYAIAR RIVDLHSRIE ESIDRVYSLD DIRRYLLFAR QFKPKISKES EDFIVEQYKH 600
601 LRQRDGSGVT KSSWRITVRQ LESMIRLSEA MARMHCCDEV QPKHVKEAFR LLNKSIIRVE 660
661 TPDVNLDQEE EIQMEVDEGA GGINGHADSP APVNGINGYN EDINQESAPK ASLRLGFSEY 720
721 CRISNLIVLH LRKVEEEEDE SALKRSELVN WYLKEIESEI DSEEELINKK RIIEKVIHRL 780
781 THYDHVLIEL TQAGLKGSTE GSESYEEDPY LVVNPNYLLE D |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
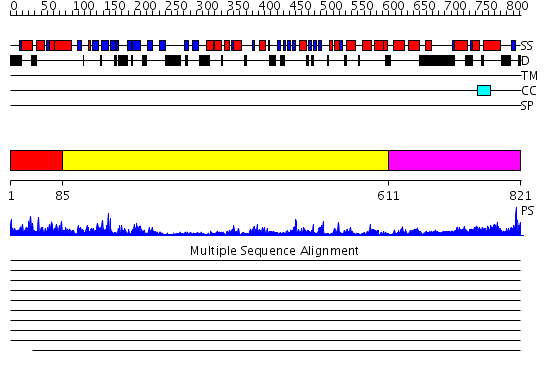
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 31.69897 | DNA replication initiator (cdc21/cdc54) N-terminal domain | |
| 2 | View Details | [85..610] | 24.39794 | N-terminal, ClpS-binding domain of ClpA, an Hsp100 chaperone; ClpA, an Hsp100 chaperone, AAA+ modules | |
| 3 | View Details | [611..821] | 24.522879 | No description for 2dhrA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)