













| General Information: |
|
| Name(s) found: |
VARS
[HGNC (HUGO)]
|
| Description(s) found:
Found 40 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 1264 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTLYVSPHP DAFPSLRALI AARYGEAGEG PGWGGAHPRI CLQPPPTSRT PFPPPRLPAL 60
61 EQGPGGLWVW GATAVAQLLW PAGLGGPGGS RAAVLVQQWV SYADTELIPA ACGATLPALG 120
121 LRSSAQDPQA VLGALGRALS PLEEWLRLHT YLAGEAPTLA DLAAVTALLL PFRYVLDPPA 180
181 RRIWNNVTRW FVTCVRQPEF RAVLGEVVLY SGARPLSHQP GPEAPALPKT AAQLKKEAKK 240
241 REKLEKFQQK QKIQQQQPPP GEKKPKPEKR EKRDPGVITY DLPTPPGEKK DVSGPMPDSY 300
301 SPRYVEAAWY PWWEQQGFFK PEYGRPNVSA ANPRGVFMMC IPPPNVTGSL HLGHALTNAI 360
361 QDSLTRWHRM RGETTLWNPG CDHAGIATQV VVEKKLWREQ GLSRHQLGRE AFLQEVWKWK 420
421 EEKGDRIYHQ LKKLGSSLDW DRACFTMDPK LSAAVTEAFV RLHEEGIIYR STRLVNWSCT 480
481 LNSAISDIEV DKKELTGRTL LSVPGYKEKV EFGVLVSFAY KVQGSDSDEE VVVATTRIET 540
541 MLGDVAVAVH PKDTRYQHLK GKNVIHPFLS RSLPIVFDEF VDMDFGTGAV KITPAHDQND 600
601 YEVGQRHGLE AISIMDSRGA LINVPPPFLG LPRFEARKAV LVALKERGLF RGIEDNPMVV 660
661 PLCNRSKDVV EPLLRPQWYV RCGEMAQAAS AAVTRGDLRI LPEAHQRTWH AWMDNIREWC 720
721 ISRQLWWGHR IPAYFVTVSD PAVPPGEDPD GRYWVSGRNE AEAREKAAKE FGVSPDKISL 780
781 QQDEDVLDTW FSSGLFPLSI LGWPNQSEDL SVFYPGTLLE TGHDILFFWV ARMVMLGLKL 840
841 TGRLPFREVY LHAIVRDAHG RKMSKSLGNV IDPLDVIYGI SLQGLHNQLL NSNLDPSEVE 900
901 KAKEGQKADF PAGIPECGTD ALRFGLCAYM SQGRDINLDV NRILGYRHFC NKLWNATKFA 960
961 LRGLGKGFVP SPTSQPGGHE SLVDRWIRSR LTEAVRLSNQ GFQAYDFPAV TTAQYSFWLY1020
1021 ELCDVYLECL KPVLNGVDQV AAECARQTLY TCLDVGLRLL SPFMPFVTEE LFQRLPRRMP1080
1081 QAPPSLCVTP YPEPSECSWK DPEAEAALEL ALSITRAVRS LRADYNLTRI RPDCFLEVAD1140
1141 EATGALASAV SGYVQALASA GVVAVLALGA PAPQGCAVAL ASDRCSIHLQ LQGLVDPARE1200
1201 LGKLQAKRVE AQRQAQRLRE RRAASGYPVK VPLEVQEADE AKLQQTEAEL RKVDEAIALF1260
1261 QKML |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
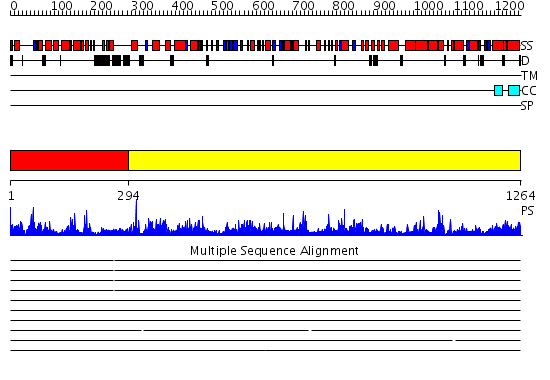
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..293] | 31.221849 | Class theta GST | |
| 2 | View Details | [294..1264] | 1000.0 | Valyl-tRNA synthetase (ValRS) C-terminal domain; Valyl-tRNA synthetase (ValRS) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.61 |
Source: Reynolds et al. (2008)